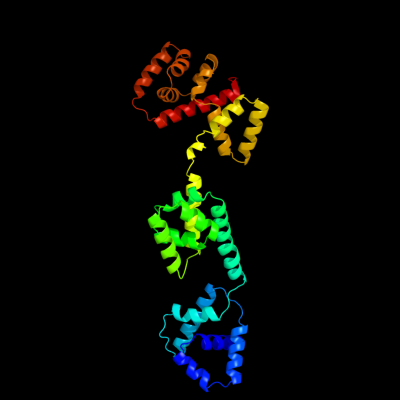
1 c3hjlA_
100.0
28
PDB header: proton transportChain: A: PDB Molecule: flagellar motor switch protein flig;PDBTitle: the structure of full-length flig from aquifex aeolicus
2 c3pl4A_
100.0
31
PDB header: motor proteinChain: A: PDB Molecule: flagellar motor switch protein;PDBTitle: crystal structure of flig (residue 116-343) from h. pylori
3 c3pkrA_
100.0
33
PDB header: motor proteinChain: A: PDB Molecule: flagellar motor switch protein;PDBTitle: crystal structure of flig (residue 86-343) from h. pylori
4 d1lkvx_
100.0
42
Fold: alpha-alpha superhelixSuperfamily: FliGFamily: FliG5 d1qc7a_
100.0
53
Fold: alpha-alpha superhelixSuperfamily: FliGFamily: FliG6 d1qc7b_
100.0
54
Fold: alpha-alpha superhelixSuperfamily: FliGFamily: FliG7 c3sohB_
99.8
29
PDB header: motor proteinChain: B: PDB Molecule: flagellar motor switch protein flig;PDBTitle: architecture of the flagellar rotor
8 d2ouxa1
98.5
15
Fold: alpha-alpha superhelixSuperfamily: MgtE N-terminal domain-likeFamily: MgtE N-terminal domain-like9 c2ouxB_
98.3
15
PDB header: transport proteinChain: B: PDB Molecule: magnesium transporter;PDBTitle: crystal structure of the soluble part of a magnesium transporter
10 d2yvxa1
98.1
14
Fold: alpha-alpha superhelixSuperfamily: MgtE N-terminal domain-likeFamily: MgtE N-terminal domain-like11 c2yvxD_
96.7
21
PDB header: transport proteinChain: D: PDB Molecule: mg2+ transporter mgte;PDBTitle: crystal structure of magnesium transporter mgte
12 c2yvzA_
96.6
17
PDB header: transport proteinChain: A: PDB Molecule: mg2+ transporter mgte;PDBTitle: crystal structure of magnesium transporter mgte cytosolic domain,2 mg2+-free form
13 c3b37A_
94.3
14
PDB header: hydrolaseChain: A: PDB Molecule: aminopeptidase n;PDBTitle: crystal structure of e. coli aminopeptidase n in complex with tyrosine
14 c2kn6A_
87.2
13
PDB header: apoptosisChain: A: PDB Molecule: apoptosis-associated speck-like protein containing a card;PDBTitle: structure of full-length human asc (apoptosis-associated speck-like2 protein containing a card)
15 c3ebhA_
78.2
8
PDB header: hydrolase inhibitorChain: A: PDB Molecule: m1 family aminopeptidase;PDBTitle: structure of the m1 alanylaminopeptidase from malaria complexed with2 bestatin
16 c1hjpA_
76.2
12
PDB header: dna recombinationChain: A: PDB Molecule: ruva;PDBTitle: holliday junction binding protein ruva from e. coli
17 c1d8lA_
72.0
13
PDB header: gene regulationChain: A: PDB Molecule: protein (holliday junction dna helicase ruva);PDBTitle: e. coli holliday junction binding protein ruva nh2 region2 lacking domain iii
18 d1cuka2
70.9
13
Fold: SAM domain-likeSuperfamily: RuvA domain 2-likeFamily: DNA helicase RuvA subunit, middle domain19 d1ixra1
56.9
16
Fold: SAM domain-likeSuperfamily: RuvA domain 2-likeFamily: DNA helicase RuvA subunit, middle domain20 c2h5xA_
56.6
12
PDB header: dna binding proteinChain: A: PDB Molecule: holliday junction atp-dependent dna helicase ruva;PDBTitle: ruva from mycobacterium tuberculosis
21 d1bvsa2
not modelled
56.3
14
Fold: SAM domain-likeSuperfamily: RuvA domain 2-likeFamily: DNA helicase RuvA subunit, middle domain22 c2x49A_
not modelled
49.4
18
PDB header: protein transportChain: A: PDB Molecule: invasion protein inva;PDBTitle: crystal structure of the c-terminal domain of inva
23 c1ixrA_
not modelled
49.4
16
PDB header: hydrolaseChain: A: PDB Molecule: holliday junction dna helicase ruva;PDBTitle: ruva-ruvb complex
24 d1bh9b_
not modelled
46.4
20
Fold: Histone-foldSuperfamily: Histone-foldFamily: TBP-associated factors, TAFs25 c3lk2B_
not modelled
38.7
14
PDB header: protein bindingChain: B: PDB Molecule: f-actin-capping protein subunit beta isoforms 1 and 2;PDBTitle: crystal structure of capz bound to the uncapping motif from carmil
26 d1iznb_
not modelled
34.2
14
Fold: Subunits of heterodimeric actin filament capping protein CapzSuperfamily: Subunits of heterodimeric actin filament capping protein CapzFamily: Capz beta-1 subunit27 d2af7a1
not modelled
33.6
18
Fold: AhpD-likeSuperfamily: AhpD-likeFamily: CMD-like28 d1szaa_
not modelled
28.0
8
Fold: alpha-alpha superhelixSuperfamily: ENTH/VHS domainFamily: RPR domain (SMART 00582 )29 c3kxrA_
not modelled
27.6
22
PDB header: transport proteinChain: A: PDB Molecule: magnesium transporter, putative;PDBTitle: structure of the cystathionine beta-synthase pair domain of the2 putative mg2+ transporter so5017 from shewanella oneidensis mr-1.
30 c1lj2B_
not modelled
26.2
21
PDB header: viral protein/ translationChain: B: PDB Molecule: nonstructural rna-binding protein 34;PDBTitle: recognition of eif4g by rotavirus nsp3 reveals a basis for2 mrna circularization
31 c2w9yA_
not modelled
24.7
9
PDB header: lipid transportChain: A: PDB Molecule: fatty acid/retinol binding protein protein 7,PDBTitle: the structure of the lipid binding protein ce-far-7 from2 caenorhabditis elegans
32 d1x2ia1
not modelled
24.2
18
Fold: SAM domain-likeSuperfamily: RuvA domain 2-likeFamily: Hef domain-like33 c2kvcA_
not modelled
23.8
18
PDB header: unknown functionChain: A: PDB Molecule: putative uncharacterized protein;PDBTitle: solution structure of the mycobacterium tuberculosis protein rv0543c,2 a member of the duf3349 superfamily. seattle structural genomics3 center for infectious disease target mytud.17112.a
34 c2lkyA_
not modelled
23.7
6
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: uncharacterized protein;PDBTitle: solution structure of msmeg_1053, the second duf3349 annotated protein2 in the genome of mycobacterium smegmatis, seattle structural genomics3 center for infectious disease target mysma.17112.b
35 d1mpga1
not modelled
23.5
13
Fold: DNA-glycosylaseSuperfamily: DNA-glycosylaseFamily: DNA repair glycosylase, 2 C-terminal domains36 d2i0za2
not modelled
23.4
16
Fold: HI0933 insert domain-likeSuperfamily: HI0933 insert domain-likeFamily: HI0933 insert domain-like37 c3thgA_
not modelled
20.8
3
PDB header: protein bindingChain: A: PDB Molecule: ribulose bisphosphate carboxylase/oxygenase activase 1,PDBTitle: crystal structure of the creosote rubisco activase c-domain
38 c3dadA_
not modelled
19.6
15
PDB header: signaling proteinChain: A: PDB Molecule: fh1/fh2 domain-containing protein 1;PDBTitle: crystal structure of the n-terminal regulatory domains of2 the formin fhod1
39 d2bgwa1
not modelled
19.3
17
Fold: SAM domain-likeSuperfamily: RuvA domain 2-likeFamily: Hef domain-like40 c8iczA_
not modelled
19.3
17
PDB header: transferase/dnaChain: A: PDB Molecule: protein (dna polymerase beta (e.c.2.7.7.7));PDBTitle: dna polymerase beta (pol b) (e.c.2.7.7.7) complexed with2 seven base pairs of dna; soaked in the presence of of datp3 (1 millimolar), mncl2 (5 millimolar), and lithium sulfate4 (75 millimolar)
41 d2peoa1
not modelled
18.9
16
Fold: RbcX-likeSuperfamily: RbcX-likeFamily: RbcX-like42 c2peoA_
not modelled
18.9
16
PDB header: chaperoneChain: A: PDB Molecule: rbcx protein;PDBTitle: crystal structure of rbcx from anabaena ca
43 c2l0kA_
not modelled
18.5
18
PDB header: transcriptionChain: A: PDB Molecule: stage iii sporulation protein d;PDBTitle: nmr solution structure of a transcription factor spoiiid in complex2 with dna
44 d2d1ha1
not modelled
18.3
11
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: TrmB-like45 c2ihmA_
not modelled
17.4
19
PDB header: transferase/dnaChain: A: PDB Molecule: dna polymerase mu;PDBTitle: polymerase mu in ternary complex with gapped 11mer dna2 duplex and bound incoming nucleotide
46 d1bh9a_
not modelled
16.9
15
Fold: Histone-foldSuperfamily: Histone-foldFamily: TBP-associated factors, TAFs47 d1nexb1
not modelled
16.7
10
Fold: F-box domainSuperfamily: F-box domainFamily: F-box domain48 d2efva1
not modelled
15.6
19
Fold: Ribbon-helix-helixSuperfamily: Ribbon-helix-helixFamily: MJ0366-like49 d2a0ma1
not modelled
15.4
15
Fold: Arginase/deacetylaseSuperfamily: Arginase/deacetylaseFamily: Arginase-like amidino hydrolases50 d1y0na_
not modelled
14.5
19
Fold: YehU-likeSuperfamily: YehU-likeFamily: YehU-like51 d1pzna1
not modelled
14.4
12
Fold: SAM domain-likeSuperfamily: Rad51 N-terminal domain-likeFamily: DNA repair protein Rad51, N-terminal domain52 d1dgna_
not modelled
14.3
18
Fold: DEATH domainSuperfamily: DEATH domainFamily: Caspase recruitment domain, CARD53 d1i5za1
not modelled
13.8
12
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: CAP C-terminal domain-like54 c2d0sA_
not modelled
13.7
0
PDB header: electron transportChain: A: PDB Molecule: cytochrome c;PDBTitle: crystal structure of the cytochrome c552 from moderate2 thermophilic bacterium, hydrogenophilus thermoluteolus
55 d1dgsa1
not modelled
13.2
20
Fold: SAM domain-likeSuperfamily: RuvA domain 2-likeFamily: NAD+-dependent DNA ligase, domain 356 c1qzeA_
not modelled
13.1
15
PDB header: replicationChain: A: PDB Molecule: uv excision repair protein rad23 homolog a;PDBTitle: hhr23a protein structure based on residual dipolar coupling2 data
57 d1cc5a_
not modelled
12.4
17
Fold: Cytochrome cSuperfamily: Cytochrome cFamily: monodomain cytochrome c58 c2jxuA_
not modelled
12.3
13
PDB header: unknown functionChain: A: PDB Molecule: terb;PDBTitle: nmr solution structure of kp-terb, a tellurite resistance2 protein from klebsiella pneumoniae
59 c2kwuA_
not modelled
12.1
9
PDB header: protein binding/signaling proteinChain: A: PDB Molecule: dna polymerase iota;PDBTitle: solution structure of ubm2 of murine polymerase iota in complex with2 ubiquitin
60 c3hjbA_
not modelled
11.9
8
PDB header: isomeraseChain: A: PDB Molecule: glucose-6-phosphate isomerase;PDBTitle: 1.5 angstrom crystal structure of glucose-6-phosphate isomerase from2 vibrio cholerae.
61 d1m8za_
not modelled
11.6
9
Fold: alpha-alpha superhelixSuperfamily: ARM repeatFamily: Pumilio repeat62 d1htaa_
not modelled
11.3
15
Fold: Histone-foldSuperfamily: Histone-foldFamily: Archaeal histone63 c2gtqA_
not modelled
11.3
16
PDB header: hydrolaseChain: A: PDB Molecule: aminopeptidase n;PDBTitle: crystal structure of aminopeptidase n from human pathogen neisseria2 meningitidis
64 d1y6xa1
not modelled
11.1
12
Fold: all-alpha NTP pyrophosphatasesSuperfamily: all-alpha NTP pyrophosphatasesFamily: HisE-like (PRA-PH)65 d1tuza_
not modelled
11.0
20
Fold: EF Hand-likeSuperfamily: EF-handFamily: EF-hand modules in multidomain proteins66 c2qezC_
not modelled
10.8
11
PDB header: lyaseChain: C: PDB Molecule: ethanolamine ammonia-lyase heavy chain;PDBTitle: crystal structure of ethanolamine ammonia-lyase heavy chain2 (yp_013784.1) from listeria monocytogenes 4b f2365 at 2.15 a3 resolution
67 d1ffgb_
not modelled
10.2
18
Fold: Ferredoxin-likeSuperfamily: CheY-binding domain of CheAFamily: CheY-binding domain of CheA68 c2krcA_
not modelled
10.2
20
PDB header: transcriptionChain: A: PDB Molecule: dna-directed rna polymerase subunit delta;PDBTitle: solution structure of the n-terminal domain of bacillus2 subtilis delta subunit of rna polymerase
69 c2l6aA_
not modelled
10.0
13
PDB header: signaling proteinChain: A: PDB Molecule: nacht, lrr and pyd domains-containing protein 12;PDBTitle: three-dimensional structure of the n-terminal effector pyrin domain of2 nlrp12
70 d2b0la1
not modelled
10.0
22
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: CodY HTH domain71 c3nbuC_
not modelled
10.0
8
PDB header: isomeraseChain: C: PDB Molecule: glucose-6-phosphate isomerase;PDBTitle: crystal structure of pgi glucosephosphate isomerase
72 d2a7wa1
not modelled
10.0
11
Fold: all-alpha NTP pyrophosphatasesSuperfamily: all-alpha NTP pyrophosphatasesFamily: HisE-like (PRA-PH)73 c2a7wF_
not modelled
10.0
11
PDB header: hydrolaseChain: F: PDB Molecule: phosphoribosyl-atp pyrophosphatase;PDBTitle: crystal structure of phosphoribosyl-atp pyrophosphatase2 from chromobacterium violaceum (atcc 12472). nesg target3 cvr7
74 c1a0oH_
not modelled
9.9
18
PDB header: chemotaxisChain: H: PDB Molecule: chea;PDBTitle: chey-binding domain of chea in complex with chey
75 c3g2bA_
not modelled
9.9
14
PDB header: biosynthetic proteinChain: A: PDB Molecule: coenzyme pqq synthesis protein d;PDBTitle: crystal structure of pqqd from xanthomonas campestris
76 d1nira1
not modelled
9.7
9
Fold: Cytochrome cSuperfamily: Cytochrome cFamily: N-terminal (heme c) domain of cytochrome cd1-nitrite reductase77 c2jmkA_
not modelled
9.7
20
PDB header: protein bindingChain: A: PDB Molecule: hypothetical protein ta0956;PDBTitle: solution structure of ta0956
78 c3ka1A_
not modelled
9.7
16
PDB header: chaperoneChain: A: PDB Molecule: rbcx protein;PDBTitle: crystal structure of rbcx from thermosynechococcus elongatus
79 d1dvha_
not modelled
9.6
28
Fold: Cytochrome cSuperfamily: Cytochrome cFamily: monodomain cytochrome c80 c2vckC_
not modelled
9.5
28
PDB header: oxidoreductaseChain: C: PDB Molecule: cyanobacterial phycoerythrobilin;PDBTitle: structure of phycoerythrobilin synthase pebs from the2 cyanophage p-ssm2 in complex with the bound substrate3 biliverdin ixa
81 d1hiob_
not modelled
9.3
15
Fold: Histone-foldSuperfamily: Histone-foldFamily: Nucleosome core histones82 d1a56a_
not modelled
9.2
11
Fold: Cytochrome cSuperfamily: Cytochrome cFamily: monodomain cytochrome c83 d1p3mh_
not modelled
9.1
17
Fold: Histone-foldSuperfamily: Histone-foldFamily: Nucleosome core histones84 c2km6A_
not modelled
9.0
4
PDB header: signaling protein, protein bindingChain: A: PDB Molecule: nacht, lrr and pyd domains-containing protein 7;PDBTitle: nmr structure of the nlrp7 pyrin domain
85 d1c53a_
not modelled
8.9
22
Fold: Cytochrome cSuperfamily: Cytochrome cFamily: monodomain cytochrome c86 c1cauB_
not modelled
8.8
11
PDB header: seed storage proteinChain: B: PDB Molecule: canavalin;PDBTitle: determination of three crystal structures of canavalin by molecular2 replacement
87 d2h6ca1
not modelled
8.6
18
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: CAP C-terminal domain-like88 d1uija1
not modelled
8.6
11
Fold: Double-stranded beta-helixSuperfamily: RmlC-like cupinsFamily: Germin/Seed storage 7S protein89 c2qwwB_
not modelled
8.5
24
PDB header: transcriptionChain: B: PDB Molecule: transcriptional regulator, marr family;PDBTitle: crystal structure of multiple antibiotic-resistance repressor (marr)2 (yp_013417.1) from listeria monocytogenes 4b f2365 at 2.07 a3 resolution
90 c1qloA_
not modelled
8.3
33
PDB header: membrane proteinsChain: A: PDB Molecule: herpes simplex virus protein icp47;PDBTitle: structure of the active domain of the herpes simplex virus2 protein icp47 in water/sodium dodecyl sulfate solution3 determined by nuclear magnetic resonance spectroscopy
91 c2k4bA_
not modelled
8.2
15
PDB header: dna binding proteinChain: A: PDB Molecule: transcriptional regulator;PDBTitle: copr repressor structure
92 d1huua_
not modelled
8.2
19
Fold: IHF-like DNA-binding proteinsSuperfamily: IHF-like DNA-binding proteinsFamily: Prokaryotic DNA-bending protein93 c2zzsW_
not modelled
8.1
22
PDB header: electron transportChain: W: PDB Molecule: PDBTitle: crystal structure of cytochrome c554 from vibrio2 parahaemolyticus strain rimd2210633
94 c1dgrW_
not modelled
8.1
7
PDB header: plant proteinChain: W: PDB Molecule: canavalin;PDBTitle: refined crystal structure of canavalin from jack bean
95 d2phla1
not modelled
8.0
19
Fold: Double-stranded beta-helixSuperfamily: RmlC-like cupinsFamily: Germin/Seed storage 7S protein96 c3fgxA_
not modelled
7.8
4
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: rbstp2171;PDBTitle: structure of uncharacterised protein rbstp2171 from bacillus2 stearothermophilus
97 d1m70a2
not modelled
7.7
22
Fold: Cytochrome cSuperfamily: Cytochrome cFamily: Two-domain cytochrome c98 d1k1va_
not modelled
7.7
22
Fold: A DNA-binding domain in eukaryotic transcription factorsSuperfamily: A DNA-binding domain in eukaryotic transcription factorsFamily: A DNA-binding domain in eukaryotic transcription factors99 d1cora_
not modelled
7.5
11
Fold: Cytochrome cSuperfamily: Cytochrome cFamily: monodomain cytochrome c