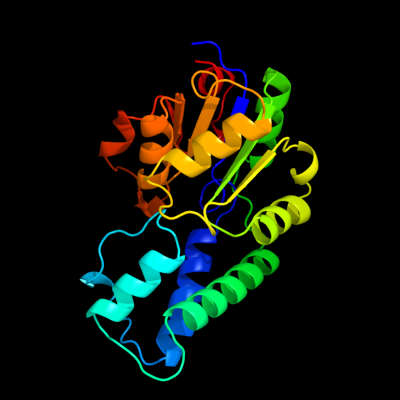
| 1 | d1te2a_
|
|
 |
100.0 |
96 |
Fold:HAD-like
Superfamily:HAD-like
Family:beta-Phosphoglucomutase-like |
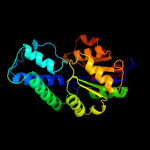
| 2 | c3iruA_
|
|
 |
100.0 |
19 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:phoshonoacetaldehyde hydrolase like protein;
PDBTitle: crystal structure of phoshonoacetaldehyde hydrolase like protein from2 oleispira antarctica
|
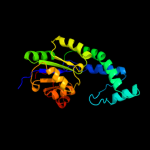
| 3 | c3d6jA_
|
|
 |
100.0 |
18 |
PDB header:hydrolase
Chain: A: PDB Molecule:putative haloacid dehalogenase-like hydrolase;
PDBTitle: crystal structure of putative haloacid dehalogenase-like hydrolase2 from bacteroides fragilis
|
| 4 | d2hsza1
|
|
 |
100.0 |
20 |
Fold:HAD-like
Superfamily:HAD-like
Family:beta-Phosphoglucomutase-like |
| 5 | d1swva_
|
|
 |
100.0 |
23 |
Fold:HAD-like
Superfamily:HAD-like
Family:Phosphonoacetaldehyde hydrolase-like |
| 6 | c3l5kA_
|
|
 |
100.0 |
24 |
PDB header:hydrolase
Chain: A: PDB Molecule:haloacid dehalogenase-like hydrolase domain-
PDBTitle: the crystal structure of human haloacid dehalogenase-like2 hydrolase domain containing 1a (hdhd1a)
|
| 7 | c3dv9A_
|
|
 |
100.0 |
20 |
PDB header:isomerase
Chain: A: PDB Molecule:beta-phosphoglucomutase;
PDBTitle: putative beta-phosphoglucomutase from bacteroides vulgatus.
|
| 8 | d2fdra1
|
|
 |
100.0 |
21 |
Fold:HAD-like
Superfamily:HAD-like
Family:beta-Phosphoglucomutase-like |
| 9 | c3e58A_
|
|
 |
100.0 |
26 |
PDB header:isomerase
Chain: A: PDB Molecule:putative beta-phosphoglucomutase;
PDBTitle: crystal structure of putative beta-phosphoglucomutase from2 streptococcus thermophilus
|
| 10 | c3mc1A_
|
|
 |
100.0 |
16 |
PDB header:hydrolase
Chain: A: PDB Molecule:predicted phosphatase, had family;
PDBTitle: crystal structure of a predicted phosphatase from2 clostridium acetobutylicum
|
| 11 | c2yy6B_
|
|
 |
100.0 |
20 |
PDB header:hydrolase
Chain: B: PDB Molecule:phosphoglycolate phosphatase;
PDBTitle: crystal structure of the phosphoglycolate phosphatase from aquifex2 aeolicus vf5
|
| 12 | c3s6jC_
|
|
 |
100.0 |
17 |
PDB header:hydrolase
Chain: C: PDB Molecule:hydrolase, haloacid dehalogenase-like family;
PDBTitle: the crystal structure of a hydrolase from pseudomonas syringae
|
| 13 | d2ah5a1
|
|
 |
100.0 |
12 |
Fold:HAD-like
Superfamily:HAD-like
Family:beta-Phosphoglucomutase-like |
| 14 | c2qltA_
|
|
 |
100.0 |
18 |
PDB header:hydrolase
Chain: A: PDB Molecule:(dl)-glycerol-3-phosphatase 1;
PDBTitle: crystal structure of an isoform of dl-glycerol-3-phosphatase, rhr2p,2 from saccharomyces cerevisiae
|
| 15 | c2hi0B_
|
|
 |
100.0 |
19 |
PDB header:hydrolase
Chain: B: PDB Molecule:putative phosphoglycolate phosphatase;
PDBTitle: crystal structure of putative phosphoglycolate phosphatase2 (yp_619066.1) from lactobacillus delbrueckii subsp. bulgaricus atcc3 baa-365 at 1.51 a resolution
|
| 16 | d2hdoa1
|
|
 |
100.0 |
18 |
Fold:HAD-like
Superfamily:HAD-like
Family:beta-Phosphoglucomutase-like |
| 17 | c3sd7A_
|
|
 |
100.0 |
16 |
PDB header:hydrolase
Chain: A: PDB Molecule:putative phosphatase;
PDBTitle: 1.7 angstrom resolution crystal structure of putative phosphatase from2 clostridium difficile
|
| 18 | c2pibA_
|
|
 |
100.0 |
27 |
PDB header:hydrolase
Chain: A: PDB Molecule:phosphorylated carbohydrates phosphatase tm_1254;
PDBTitle: crystal structure of putative beta-phosphoglucomutase from2 thermotoga maritima
|
| 19 | d2go7a1
|
|
 |
99.9 |
16 |
Fold:HAD-like
Superfamily:HAD-like
Family:beta-Phosphoglucomutase-like |
| 20 | c3nuqA_
|
|
 |
99.9 |
13 |
PDB header:hydrolase
Chain: A: PDB Molecule:putative nucleotide phosphatase;
PDBTitle: structure of a putative nucleotide phosphatase from saccharomyces2 cerevisiae
|
| 21 | c2no5B_ |
|
not modelled |
99.9 |
20 |
PDB header:hydrolase
Chain: B: PDB Molecule:(s)-2-haloacid dehalogenase iva;
PDBTitle: crystal structure analysis of a dehalogenase with intermediate complex
|
| 22 | d1zs9a1 |
|
not modelled |
99.9 |
15 |
Fold:HAD-like
Superfamily:HAD-like
Family:Enolase-phosphatase E1 |
| 23 | d1zrna_ |
|
not modelled |
99.9 |
16 |
Fold:HAD-like
Superfamily:HAD-like
Family:HAD-related |
| 24 | c3qnmA_ |
|
not modelled |
99.9 |
13 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:haloacid dehalogenase-like hydrolase;
PDBTitle: haloalkane dehalogenase family member from bacteroides2 thetaiotaomicron of unknown function
|
| 25 | c2om6A_ |
|
not modelled |
99.9 |
17 |
PDB header:hydrolase
Chain: A: PDB Molecule:probable phosphoserine phosphatase;
PDBTitle: hypothetical protein (probable phosphoserine phosph (ph0253) from2 pyrococcus horikoshii ot3
|
| 26 | d1o08a_ |
|
not modelled |
99.9 |
22 |
Fold:HAD-like
Superfamily:HAD-like
Family:beta-Phosphoglucomutase-like |
| 27 | c3ddhA_ |
|
not modelled |
99.9 |
22 |
PDB header:hydrolase
Chain: A: PDB Molecule:putative haloacid dehalogenase-like family hydrolase;
PDBTitle: the structure of a putative haloacid dehalogenase-like family2 hydrolase from bacteroides thetaiotaomicron vpi-5482
|
| 28 | c3m9lA_ |
|
not modelled |
99.9 |
20 |
PDB header:hydrolase
Chain: A: PDB Molecule:hydrolase, haloacid dehalogenase-like family;
PDBTitle: crystal structure of probable had family hydrolase from2 pseudomonas fluorescens pf-5
|
| 29 | c3ed5A_ |
|
not modelled |
99.9 |
13 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:yfnb;
PDBTitle: the crystal structure of yfnb from bacillus subtilis subsp. subtilis2 str. 168
|
| 30 | d2hcfa1 |
|
not modelled |
99.9 |
19 |
Fold:HAD-like
Superfamily:HAD-like
Family:beta-Phosphoglucomutase-like |
| 31 | c2hoqA_ |
|
not modelled |
99.9 |
15 |
PDB header:hydrolase
Chain: A: PDB Molecule:putative had-hydrolase ph1655;
PDBTitle: crystal structure of the probable haloacid dehalogenase (ph1655) from2 pyrococcus horikoshii ot3
|
| 32 | d2gfha1 |
|
not modelled |
99.9 |
19 |
Fold:HAD-like
Superfamily:HAD-like
Family:beta-Phosphoglucomutase-like |
| 33 | d1x42a1 |
|
not modelled |
99.9 |
22 |
Fold:HAD-like
Superfamily:HAD-like
Family:HAD-related |
| 34 | c3nasA_ |
|
not modelled |
99.9 |
23 |
PDB header:isomerase
Chain: A: PDB Molecule:beta-phosphoglucomutase;
PDBTitle: the crystal structure of beta-phosphoglucomutase from bacillus2 subtilis
|
| 35 | d1qq5a_ |
|
not modelled |
99.9 |
19 |
Fold:HAD-like
Superfamily:HAD-like
Family:HAD-related |
| 36 | c2w11B_ |
|
not modelled |
99.9 |
18 |
PDB header:hydrolase
Chain: B: PDB Molecule:2-haloalkanoic acid dehalogenase;
PDBTitle: structure of the l-2-haloacid dehalogenase from sulfolobus2 tokodaii
|
| 37 | c2g80C_ |
|
not modelled |
99.9 |
11 |
PDB header:hydrolase
Chain: C: PDB Molecule:protein utr4;
PDBTitle: crystal structure of utr4 protein (unknown transcript 4 protein)2 (yel038w) from saccharomyces cerevisiae at 2.28 a resolution
|
| 38 | d2g80a1 |
|
not modelled |
99.9 |
11 |
Fold:HAD-like
Superfamily:HAD-like
Family:Enolase-phosphatase E1 |
| 39 | c3k1zA_ |
|
not modelled |
99.9 |
18 |
PDB header:hydrolase
Chain: A: PDB Molecule:haloacid dehalogenase-like hydrolase domain-containing
PDBTitle: crystal structure of human haloacid dehalogenase-like hydrolase domain2 containing 3 (hdhd3)
|
| 40 | c2pkeA_ |
|
not modelled |
99.9 |
20 |
PDB header:hydrolase
Chain: A: PDB Molecule:haloacid delahogenase-like family hydrolase;
PDBTitle: crystal structure of haloacid delahogenase-like family hydrolase2 (np_639141.1) from xanthomonas campestris at 1.81 a resolution
|
| 41 | c3cnhA_ |
|
not modelled |
99.9 |
15 |
PDB header:hydrolase
Chain: A: PDB Molecule:hydrolase family protein;
PDBTitle: crystal structure of predicted hydrolase of haloacid dehalogenase-like2 superfamily (np_295428.1) from deinococcus radiodurans at 1.66 a3 resolution
|
| 42 | d2fi1a1 |
|
not modelled |
99.9 |
16 |
Fold:HAD-like
Superfamily:HAD-like
Family:Phosphonoacetaldehyde hydrolase-like |
| 43 | c3kd3A_ |
|
not modelled |
99.9 |
13 |
PDB header:unknown function
Chain: A: PDB Molecule:phosphoserine phosphohydrolase-like protein;
PDBTitle: crystal structure of a phosphoserine phosphohydrolase-like protein2 from francisella tularensis subsp. tularensis schu s4
|
| 44 | c3kzxA_ |
|
not modelled |
99.9 |
17 |
PDB header:hydrolase
Chain: A: PDB Molecule:had-superfamily hydrolase, subfamily ia, variant 1;
PDBTitle: crystal structure of a had-superfamily hydrolase from ehrlichia2 chaffeensis at 1.9a resolution
|
| 45 | c2ho4A_ |
|
not modelled |
99.9 |
13 |
PDB header:hydrolase
Chain: A: PDB Molecule:haloacid dehalogenase-like hydrolase domain
PDBTitle: crystal structure of protein from mouse mm.236127
|
| 46 | d2c4na1 |
|
not modelled |
99.9 |
16 |
Fold:HAD-like
Superfamily:HAD-like
Family:NagD-like |
| 47 | c3pdwA_ |
|
not modelled |
99.8 |
13 |
PDB header:hydrolase
Chain: A: PDB Molecule:uncharacterized hydrolase yutf;
PDBTitle: crystal structure of putative p-nitrophenyl phosphatase from bacillus2 subtilis
|
| 48 | c3qgmC_ |
|
not modelled |
99.8 |
17 |
PDB header:hydrolase
Chain: C: PDB Molecule:p-nitrophenyl phosphatase (pho2);
PDBTitle: p-nitrophenyl phosphatase from archaeoglobus fulgidus
|
| 49 | c3l8hC_ |
|
not modelled |
99.8 |
20 |
PDB header:hydrolase
Chain: C: PDB Molecule:putative haloacid dehalogenase-like hydrolase;
PDBTitle: crystal structure of d,d-heptose 1.7-bisphosphate phosphatase from b.2 bronchiseptica complexed with magnesium and phosphate
|
| 50 | c2p11A_ |
|
not modelled |
99.8 |
15 |
PDB header:hydrolase
Chain: A: PDB Molecule:hypothetical protein;
PDBTitle: crystal structure of a putative haloacid dehalogenase-like hydrolase2 (bxe_b1342) from burkholderia xenovorans lb400 at 2.20 a resolution
|
| 51 | d2o2xa1 |
|
not modelled |
99.8 |
21 |
Fold:HAD-like
Superfamily:HAD-like
Family:Histidinol phosphatase-like |
| 52 | d2gmwa1 |
|
not modelled |
99.8 |
23 |
Fold:HAD-like
Superfamily:HAD-like
Family:Histidinol phosphatase-like |
| 53 | d1zd3a1 |
|
not modelled |
99.8 |
16 |
Fold:HAD-like
Superfamily:HAD-like
Family:YihX-like |
| 54 | d1wvia_ |
|
not modelled |
99.8 |
12 |
Fold:HAD-like
Superfamily:HAD-like
Family:NagD-like |
| 55 | c3esqA_ |
|
not modelled |
99.8 |
23 |
PDB header:hydrolase
Chain: A: PDB Molecule:d,d-heptose 1,7-bisphosphate phosphatase;
PDBTitle: crystal structure of calcium-bound d,d-heptose 1.7-2 bisphosphate phosphatase from e. coli
|
| 56 | d1ys9a1 |
|
not modelled |
99.8 |
13 |
Fold:HAD-like
Superfamily:HAD-like
Family:NagD-like |
| 57 | c2x4dB_ |
|
not modelled |
99.8 |
15 |
PDB header:hydrolase
Chain: B: PDB Molecule:phospholysine phosphohistidine inorganic pyrophosphate
PDBTitle: crystal structure of human phospholysine phosphohistidine2 inorganic pyrophosphate phosphatase lhpp
|
| 58 | c2i6xA_ |
|
not modelled |
99.8 |
12 |
PDB header:hydrolase
Chain: A: PDB Molecule:hydrolase, haloacid dehalogenase-like family;
PDBTitle: the structure of a predicted had-like family hydrolase from2 porphyromonas gingivalis.
|
| 59 | d1ydfa1 |
|
not modelled |
99.8 |
12 |
Fold:HAD-like
Superfamily:HAD-like
Family:NagD-like |
| 60 | d1qyia_ |
|
not modelled |
99.8 |
23 |
Fold:HAD-like
Superfamily:HAD-like
Family:Hypothetical protein MW1667 (SA1546) |
| 61 | d1yv9a1 |
|
not modelled |
99.8 |
13 |
Fold:HAD-like
Superfamily:HAD-like
Family:NagD-like |
| 62 | d1vjra_ |
|
not modelled |
99.8 |
16 |
Fold:HAD-like
Superfamily:HAD-like
Family:NagD-like |
| 63 | c2cftA_ |
|
not modelled |
99.8 |
18 |
PDB header:phosphatase
Chain: A: PDB Molecule:pyridoxal phosphate phosphatase;
PDBTitle: crystal structure of human pyridoxal 5'-phosphate2 phosphatase with its substrate
|
| 64 | c2zg6A_ |
|
not modelled |
99.8 |
15 |
PDB header:hydrolase
Chain: A: PDB Molecule:putative uncharacterized protein st2620;
PDBTitle: crystal structure of hypothetical protein; probable 2-2 haloalkanoic acid dehalogenase from sulfolobus tokodaii
|
| 65 | d2b0ca1 |
|
not modelled |
99.8 |
16 |
Fold:HAD-like
Superfamily:HAD-like
Family:YihX-like |
| 66 | c1cr6A_ |
|
not modelled |
99.8 |
22 |
PDB header:hydrolase
Chain: A: PDB Molecule:epoxide hydrolase;
PDBTitle: crystal structure of murine soluble epoxide hydrolase2 complexed with cpu inhibitor
|
| 67 | d1u7pa_ |
|
not modelled |
99.8 |
17 |
Fold:HAD-like
Superfamily:HAD-like
Family:Magnesium-dependent phosphatase-1, Mdp1 |
| 68 | d1nnla_ |
|
not modelled |
99.7 |
19 |
Fold:HAD-like
Superfamily:HAD-like
Family:Phosphoserine phosphatase |
| 69 | c1zjjA_ |
|
not modelled |
99.7 |
18 |
PDB header:hydrolase
Chain: A: PDB Molecule:hypothetical protein ph1952;
PDBTitle: crystal structure of hypothetical protein ph1952 from pyrococcus2 horikoshii ot3
|
| 70 | c3m1yA_ |
|
not modelled |
99.7 |
18 |
PDB header:hydrolase
Chain: A: PDB Molecule:phosphoserine phosphatase (serb);
PDBTitle: crystal structure of a phosphoserine phosphatase (serb) from2 helicobacter pylori
|
| 71 | d1cr6a1 |
|
not modelled |
99.7 |
22 |
Fold:HAD-like
Superfamily:HAD-like
Family:YihX-like |
| 72 | d1j97a_ |
|
not modelled |
99.7 |
16 |
Fold:HAD-like
Superfamily:HAD-like
Family:Phosphoserine phosphatase |
| 73 | c2odaB_ |
|
not modelled |
99.7 |
17 |
PDB header:protein binding
Chain: B: PDB Molecule:hypothetical protein pspto_2114;
PDBTitle: crystal structure of pspto_2114
|
| 74 | d2feaa1 |
|
not modelled |
99.7 |
15 |
Fold:HAD-like
Superfamily:HAD-like
Family:MtnX-like |
| 75 | c2hx1D_ |
|
not modelled |
99.7 |
14 |
PDB header:hydrolase
Chain: D: PDB Molecule:predicted sugar phosphatases of the had
PDBTitle: crystal structure of possible sugar phosphatase, had2 superfamily (zp_00311070.1) from cytophaga hutchinsonii3 atcc 33406 at 2.10 a resolution
|
| 76 | d2fpwa1 |
|
not modelled |
99.6 |
16 |
Fold:HAD-like
Superfamily:HAD-like
Family:Histidinol phosphatase-like |
| 77 | c3ib6B_ |
|
not modelled |
99.6 |
20 |
PDB header:structural genomics, unknown function
Chain: B: PDB Molecule:uncharacterized protein;
PDBTitle: crystal structure of an uncharacterized protein from listeria2 monocytogenes serotype 4b
|
| 78 | d1rkua_ |
|
not modelled |
99.5 |
12 |
Fold:HAD-like
Superfamily:HAD-like
Family:Homoserine kinase ThrH |
| 79 | c2pr7A_ |
|
not modelled |
99.4 |
14 |
PDB header:hydrolase
Chain: A: PDB Molecule:haloacid dehalogenase/epoxide hydrolase family;
PDBTitle: crystal structure of uncharacterized protein (np_599989.1) from2 corynebacterium glutamicum atcc 13032 kitasato at 1.44 a resolution
|
| 80 | d1wr8a_ |
|
not modelled |
99.3 |
13 |
Fold:HAD-like
Superfamily:HAD-like
Family:Predicted hydrolases Cof |
| 81 | c3p96A_ |
|
not modelled |
99.3 |
20 |
PDB header:hydrolase
Chain: A: PDB Molecule:phosphoserine phosphatase serb;
PDBTitle: crystal structure of phosphoserine phosphatase serb from mycobacterium2 avium, native form
|
| 82 | d1l6ra_ |
|
not modelled |
99.3 |
14 |
Fold:HAD-like
Superfamily:HAD-like
Family:Predicted hydrolases Cof |
| 83 | d1q92a_ |
|
not modelled |
99.3 |
16 |
Fold:HAD-like
Superfamily:HAD-like
Family:5'(3')-deoxyribonucleotidase (dNT-2) |
| 84 | c2i7dB_ |
|
not modelled |
99.3 |
14 |
PDB header:hydrolase
Chain: B: PDB Molecule:5'(3')-deoxyribonucleotidase, cytosolic type;
PDBTitle: structure of human cytosolic deoxyribonucleotidase in2 complex with deoxyuridine, alf4 and mg2+
|
| 85 | c3mn1B_ |
|
not modelled |
99.2 |
20 |
PDB header:hydrolase
Chain: B: PDB Molecule:probable yrbi family phosphatase;
PDBTitle: crystal structure of probable yrbi family phosphatase from pseudomonas2 syringae pv.phaseolica 1448a
|
| 86 | d1k1ea_ |
|
not modelled |
99.2 |
19 |
Fold:HAD-like
Superfamily:HAD-like
Family:Probable phosphatase YrbI |
| 87 | c3e8mD_ |
|
not modelled |
99.2 |
18 |
PDB header:transferase
Chain: D: PDB Molecule:acylneuraminate cytidylyltransferase;
PDBTitle: structure-function analysis of 2-keto-3-deoxy-d-glycero-d-galacto-2 nononate-9-phosphate (kdn) phosphatase defines a new clad within the3 type c0 had subfamily
|
| 88 | c2r8zC_ |
|
not modelled |
99.2 |
21 |
PDB header:hydrolase
Chain: C: PDB Molecule:3-deoxy-d-manno-octulosonate 8-phosphate phosphatase;
PDBTitle: crystal structure of yrbi phosphatase from escherichia coli in complex2 with a phosphate and a calcium ion
|
| 89 | c3n07B_ |
|
not modelled |
99.1 |
20 |
PDB header:hydrolase
Chain: B: PDB Molecule:3-deoxy-d-manno-octulosonate 8-phosphate phosphatase;
PDBTitle: structure of putative 3-deoxy-d-manno-octulosonate 8-phosphate2 phosphatase from vibrio cholerae
|
| 90 | c2p9jH_ |
|
not modelled |
99.1 |
20 |
PDB header:structural genomics, unknown function
Chain: H: PDB Molecule:hypothetical protein aq2171;
PDBTitle: crystal structure of aq2171 from aquifex aeolicus
|
| 91 | c3n1uA_ |
|
not modelled |
99.1 |
19 |
PDB header:hydrolase
Chain: A: PDB Molecule:hydrolase, had superfamily, subfamily iii a;
PDBTitle: structure of putative had superfamily (subfamily iii a) hydrolase from2 legionella pneumophila
|
| 92 | c3n28A_ |
|
not modelled |
99.1 |
19 |
PDB header:hydrolase
Chain: A: PDB Molecule:phosphoserine phosphatase;
PDBTitle: crystal structure of probable phosphoserine phosphatase from vibrio2 cholerae, unliganded form
|
| 93 | c3mmzA_ |
|
not modelled |
99.0 |
19 |
PDB header:hydrolase
Chain: A: PDB Molecule:putative had family hydrolase;
PDBTitle: crystal structure of putative had family hydrolase from streptomyces2 avermitilis ma-4680
|
| 94 | c3fvvA_ |
|
not modelled |
99.0 |
14 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:uncharacterized protein;
PDBTitle: the crystal structure of the protein with unknown function from2 bordetella pertussis tohama i
|
| 95 | c3ewiB_ |
|
not modelled |
99.0 |
16 |
PDB header:transferase
Chain: B: PDB Molecule:n-acylneuraminate cytidylyltransferase;
PDBTitle: structural analysis of the c-terminal domain of murine cmp-2 sialic acid synthetase
|
| 96 | c3kc2A_ |
|
not modelled |
98.9 |
13 |
PDB header:hydrolase
Chain: A: PDB Molecule:uncharacterized protein ykr070w;
PDBTitle: crystal structure of mitochondrial had-like phosphatase from2 saccharomyces cerevisiae
|
| 97 | c2qyhD_ |
|
not modelled |
98.8 |
15 |
PDB header:structural genomics, unknown function
Chain: D: PDB Molecule:hypothetical conserved protein, gk1056;
PDBTitle: crystal structure of the hypothetical protein (gk1056) from2 geobacillus kaustophilus hta426
|
| 98 | c3fzqA_ |
|
not modelled |
98.8 |
13 |
PDB header:hydrolase
Chain: A: PDB Molecule:putative hydrolase;
PDBTitle: crystal structure of putative haloacid dehalogenase-like hydrolase2 (yp_001086940.1) from clostridium difficile 630 at 2.10 a resolution
|
| 99 | c3r4cA_ |
|
not modelled |
98.8 |
14 |
PDB header:hydrolase
Chain: A: PDB Molecule:hydrolase, haloacid dehalogenase-like hydrolase;
PDBTitle: divergence of structure and function among phosphatases of the2 haloalkanoate (had) enzyme superfamily: analysis of bt1666 from3 bacteroides thetaiotaomicron
|
| 100 | c3niwA_ |
|
not modelled |
98.7 |
13 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:haloacid dehalogenase-like hydrolase;
PDBTitle: crystal structure of a haloacid dehalogenase-like hydrolase from2 bacteroides thetaiotaomicron
|
| 101 | d2b30a1 |
|
not modelled |
98.7 |
13 |
Fold:HAD-like
Superfamily:HAD-like
Family:Predicted hydrolases Cof |
| 102 | c3l7yA_ |
|
not modelled |
98.7 |
12 |
PDB header:hydrolase
Chain: A: PDB Molecule:putative uncharacterized protein smu.1108c;
PDBTitle: the crystal structure of smu.1108c from streptococcus mutans ua159
|
| 103 | d1nf2a_ |
|
not modelled |
98.6 |
12 |
Fold:HAD-like
Superfamily:HAD-like
Family:Predicted hydrolases Cof |
| 104 | d1rkqa_ |
|
not modelled |
98.6 |
14 |
Fold:HAD-like
Superfamily:HAD-like
Family:Predicted hydrolases Cof |
| 105 | d2rbka1 |
|
not modelled |
98.6 |
11 |
Fold:HAD-like
Superfamily:HAD-like
Family:Predicted hydrolases Cof |
| 106 | d1nrwa_ |
|
not modelled |
98.6 |
16 |
Fold:HAD-like
Superfamily:HAD-like
Family:Predicted hydrolases Cof |
| 107 | d1rlma_ |
|
not modelled |
98.6 |
14 |
Fold:HAD-like
Superfamily:HAD-like
Family:Predicted hydrolases Cof |
| 108 | d1s2oa1 |
|
not modelled |
98.5 |
15 |
Fold:HAD-like
Superfamily:HAD-like
Family:Predicted hydrolases Cof |
| 109 | c3dnpA_ |
|
not modelled |
98.5 |
11 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:stress response protein yhax;
PDBTitle: crystal structure of stress response protein yhax from bacillus2 subtilis
|
| 110 | c3daoB_ |
|
not modelled |
98.4 |
12 |
PDB header:hydrolase
Chain: B: PDB Molecule:putative phosphatse;
PDBTitle: crystal structure of a putative phosphatse (eubrec_1417) from2 eubacterium rectale at 1.80 a resolution
|
| 111 | d2vkqa1 |
|
not modelled |
98.4 |
17 |
Fold:HAD-like
Superfamily:HAD-like
Family:Pyrimidine 5'-nucleotidase (UMPH-1) |
| 112 | d1yj5a1 |
|
not modelled |
98.4 |
20 |
Fold:HAD-like
Superfamily:HAD-like
Family:phosphatase domain of polynucleotide kinase |
| 113 | c3zvmA_ |
|
not modelled |
98.4 |
21 |
PDB header:hydrolase/transferase/dna
Chain: A: PDB Molecule:bifunctional polynucleotide phosphatase/kinase;
PDBTitle: the structural basis for substrate recognition by mammalian2 polynucleotide kinase 3' phosphatase
|
| 114 | c3gygA_ |
|
not modelled |
98.4 |
12 |
PDB header:hydrolase
Chain: A: PDB Molecule:ntd biosynthesis operon putative hydrolase ntdb;
PDBTitle: crystal structure of yhjk (haloacid dehalogenase-like hydrolase2 protein) from bacillus subtilis
|
| 115 | c2iyeC_ |
|
not modelled |
98.4 |
19 |
PDB header:hydrolase
Chain: C: PDB Molecule:copper-transporting atpase;
PDBTitle: structure of catalytic cpx-atpase domain copb-b
|
| 116 | d1ltqa1 |
|
not modelled |
98.4 |
21 |
Fold:HAD-like
Superfamily:HAD-like
Family:phosphatase domain of polynucleotide kinase |
| 117 | c3pgvB_ |
|
not modelled |
98.3 |
16 |
PDB header:hydrolase
Chain: B: PDB Molecule:haloacid dehalogenase-like hydrolase;
PDBTitle: crystal structure of a haloacid dehalogenase-like hydrolase2 (kpn_04322) from klebsiella pneumoniae subsp. pneumoniae mgh 78578 at3 2.39 a resolution
|
| 118 | d1wzca1 |
|
not modelled |
98.3 |
7 |
Fold:HAD-like
Superfamily:HAD-like
Family:Predicted hydrolases Cof |
| 119 | d1z5ga1 |
|
not modelled |
98.2 |
14 |
Fold:HAD-like
Superfamily:HAD-like
Family:Class B acid phosphatase, AphA |
| 120 | c1xviA_ |
|
not modelled |
98.1 |
15 |
PDB header:hydrolase
Chain: A: PDB Molecule:putative mannosyl-3-phosphoglycerate phosphatase;
PDBTitle: crystal structure of yedp, phosphatase-like domain protein2 from escherichia coli k12
|