| 1 | c1i8dB_
|
|
 |
100.0 |
100 |
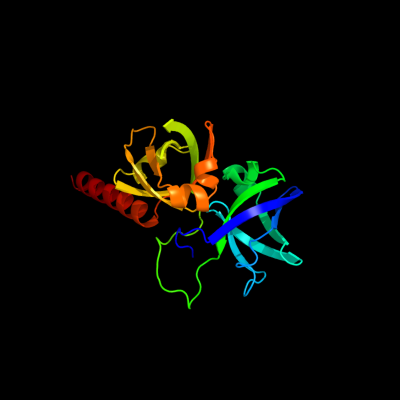
PDB header:transferase
Chain: B: PDB Molecule:riboflavin synthase;
PDBTitle: crystal structure of riboflavin synthase
|
| 2 | c1kzlA_
|
|
 |
100.0 |
35 |
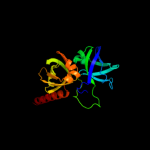
PDB header:transferase
Chain: A: PDB Molecule:riboflavin synthase;
PDBTitle: riboflavin synthase from s.pombe bound to2 carboxyethyllumazine
|
| 3 | c3a35B_
|
|
 |
100.0 |
31 |
PDB header:luminescent protein
Chain: B: PDB Molecule:lumazine protein;
PDBTitle: crystal structure of lump complexed with riboflavin
|
| 4 | c3ddyA_
|
|
 |
100.0 |
32 |
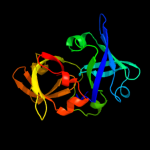
PDB header:luminescent protein
Chain: A: PDB Molecule:lumazine protein;
PDBTitle: structure of lumazine protein, an optical transponder of luminescent2 bacteria
|
| 5 | d1i8da2
|
|
 |
100.0 |
100 |
Fold:Reductase/isomerase/elongation factor common domain
Superfamily:Riboflavin synthase domain-like
Family:Riboflavin synthase |
| 6 | d1kzla2
|
|
 |
100.0 |
33 |
Fold:Reductase/isomerase/elongation factor common domain
Superfamily:Riboflavin synthase domain-like
Family:Riboflavin synthase |
| 7 | c1hzeB_
|
|
 |
100.0 |
100 |
PDB header:transferase
Chain: B: PDB Molecule:riboflavin synthase alpha chain;
PDBTitle: solution structure of the n-terminal domain of riboflavin synthase2 from e. coli
|
| 8 | c1i18B_
|
|
 |
100.0 |
100 |
PDB header:transferase
Chain: B: PDB Molecule:riboflavin synthase alpha chain;
PDBTitle: solution structure of the n-terminal domain of riboflavin synthase2 from e. coli
|
| 9 | d1i8da1
|
|
 |
100.0 |
100 |
Fold:Reductase/isomerase/elongation factor common domain
Superfamily:Riboflavin synthase domain-like
Family:Riboflavin synthase |
| 10 | c1pkvA_
|
|
 |
100.0 |
100 |
PDB header:transferase
Chain: A: PDB Molecule:riboflavin synthase alpha chain;
PDBTitle: the n-terminal domain of riboflavin synthase in complex with2 riboflavin
|
| 11 | c1pkvB_
|
|
 |
100.0 |
100 |
PDB header:transferase
Chain: B: PDB Molecule:riboflavin synthase alpha chain;
PDBTitle: the n-terminal domain of riboflavin synthase in complex with2 riboflavin
|
| 12 | d1kzla1
|
|
 |
100.0 |
38 |
Fold:Reductase/isomerase/elongation factor common domain
Superfamily:Riboflavin synthase domain-like
Family:Riboflavin synthase |
| 13 | d2g50a1
|
|
 |
68.8 |
16 |
Fold:PK beta-barrel domain-like
Superfamily:PK beta-barrel domain-like
Family:Pyruvate kinase beta-barrel domain |
| 14 | c3nrfA_
|
|
 |
64.0 |
26 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:apag protein;
PDBTitle: crystal structure of an apag protein (pa1934) from pseudomonas2 aeruginosa pao1 at 1.50 a resolution
|
| 15 | d1pkla1
|
|
 |
48.6 |
11 |
Fold:PK beta-barrel domain-like
Superfamily:PK beta-barrel domain-like
Family:Pyruvate kinase beta-barrel domain |
| 16 | d1e0ta1
|
|
 |
46.2 |
31 |
Fold:PK beta-barrel domain-like
Superfamily:PK beta-barrel domain-like
Family:Pyruvate kinase beta-barrel domain |
| 17 | d1liua1
|
|
 |
43.7 |
22 |
Fold:PK beta-barrel domain-like
Superfamily:PK beta-barrel domain-like
Family:Pyruvate kinase beta-barrel domain |
| 18 | d1a3xa1
|
|
 |
34.7 |
15 |
Fold:PK beta-barrel domain-like
Superfamily:PK beta-barrel domain-like
Family:Pyruvate kinase beta-barrel domain |
| 19 | c3o44G_
|
|
 |
33.8 |
36 |
PDB header:toxin
Chain: G: PDB Molecule:hemolysin;
PDBTitle: crystal structure of the vibrio cholerae cytolysin (hlya) heptameric2 pore
|
| 20 | d2hx0a1
|
|
 |
30.6 |
17 |
Fold:AF0104/ALDC/Ptd012-like
Superfamily:AF0104/ALDC/Ptd012-like
Family:AF0104-like |
| 21 | d1pkma1 |
|
not modelled |
29.2 |
20 |
Fold:PK beta-barrel domain-like
Superfamily:PK beta-barrel domain-like
Family:Pyruvate kinase beta-barrel domain |
| 22 | c1xezA_ |
|
not modelled |
28.2 |
36 |
PDB header:toxin
Chain: A: PDB Molecule:hemolysin;
PDBTitle: crystal structure of the vibrio cholerae cytolysin (hlya)2 pro-toxin with octylglucoside bound
|
| 23 | c1hn0A_ |
|
not modelled |
27.4 |
14 |
PDB header:lyase
Chain: A: PDB Molecule:chondroitin abc lyase i;
PDBTitle: crystal structure of chondroitin abc lyase i from proteus2 vulgaris at 1.9 angstroms resolution
|
| 24 | d1zbfa1 |
|
not modelled |
26.8 |
10 |
Fold:Ribonuclease H-like motif
Superfamily:Ribonuclease H-like
Family:Ribonuclease H |
| 25 | d3bzka4 |
|
not modelled |
25.2 |
16 |
Fold:OB-fold
Superfamily:Nucleic acid-binding proteins
Family:Cold shock DNA-binding domain-like |
| 26 | d1hn0a3 |
|
not modelled |
23.5 |
11 |
Fold:Hyaluronate lyase-like, C-terminal domain
Superfamily:Hyaluronate lyase-like, C-terminal domain
Family:Hyaluronate lyase-like, C-terminal domain |
| 27 | c1v0eB_ |
|
not modelled |
20.2 |
26 |
PDB header:hydrolase
Chain: B: PDB Molecule:endo-alpha-sialidase;
PDBTitle: endosialidase of bacteriophage k1f
|
| 28 | d1zrua1 |
|
not modelled |
17.9 |
24 |
Fold:Virus attachment protein globular domain
Superfamily:Virus attachment protein globular domain
Family:Lactophage receptor-binding protein head domain |
| 29 | c2izpB_ |
|
not modelled |
16.9 |
25 |
PDB header:toxin
Chain: B: PDB Molecule:putative membrane antigen;
PDBTitle: bipd - an invasion prtein associated with the type-iii2 secretion system of burkholderia pseudomallei.
|
| 30 | d2izpa1 |
|
not modelled |
15.4 |
25 |
Fold:IpaD-like
Superfamily:IpaD-like
Family:IpaD-like |
| 31 | d1vqoq1 |
|
not modelled |
15.2 |
24 |
Fold:SH3-like barrel
Superfamily:Translation proteins SH3-like domain
Family:Ribosomal proteins L24p and L21e |
| 32 | c3da0C_ |
|
not modelled |
13.8 |
24 |
PDB header:viral protein
Chain: C: PDB Molecule:cleaved chimeric receptor binding protein from
PDBTitle: crystal structure of a cleaved form of a chimeric receptor binding2 protein from lactococcal phages subspecies tp901-1 and p2
|
| 33 | c2khiA_ |
|
not modelled |
13.7 |
10 |
PDB header:ribosomal protein
Chain: A: PDB Molecule:30s ribosomal protein s1;
PDBTitle: nmr structure of the domain 4 of the e. coli ribosomal2 protein s1
|
| 34 | c3h43F_ |
|
not modelled |
13.2 |
20 |
PDB header:hydrolase
Chain: F: PDB Molecule:proteasome-activating nucleotidase;
PDBTitle: n-terminal domain of the proteasome-activating nucleotidase2 of methanocaldococcus jannaschii
|
| 35 | c2zkrq_ |
|
not modelled |
12.8 |
29 |
PDB header:ribosomal protein/rna
Chain: Q: PDB Molecule:rna expansion segment es31 part ii;
PDBTitle: structure of a mammalian ribosomal 60s subunit within an2 80s complex obtained by docking homology models of the rna3 and proteins into an 8.7 a cryo-em map
|
| 36 | d1qfja1 |
|
not modelled |
12.8 |
7 |
Fold:Reductase/isomerase/elongation factor common domain
Superfamily:Riboflavin synthase domain-like
Family:Ferredoxin reductase FAD-binding domain-like |
| 37 | d1xpna_ |
|
not modelled |
12.7 |
25 |
Fold:Prealbumin-like
Superfamily:Hypothetical protein PA1324
Family:Hypothetical protein PA1324 |
| 38 | c2jz2A_ |
|
not modelled |
12.4 |
44 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:ssl0352 protein;
PDBTitle: solution nmr structure of ssl0352 protein from synechocystis sp. pcc2 6803. northeast structural genomics consortium target sgr42
|
| 39 | d1xsza2 |
|
not modelled |
11.9 |
25 |
Fold:TBP-like
Superfamily:RalF, C-terminal domain
Family:RalF, C-terminal domain |
| 40 | d1yq2a3 |
|
not modelled |
11.6 |
21 |
Fold:Galactose-binding domain-like
Superfamily:Galactose-binding domain-like
Family:beta-Galactosidase/glucuronidase, N-terminal domain |
| 41 | c3kf8D_ |
|
not modelled |
11.4 |
27 |
PDB header:structural protein
Chain: D: PDB Molecule:protein ten1;
PDBTitle: crystal structure of c. tropicalis stn1-ten1 complex
|
| 42 | c1s1iQ_ |
|
not modelled |
11.2 |
35 |
PDB header:ribosome
Chain: Q: PDB Molecule:60s ribosomal protein l21-a;
PDBTitle: structure of the ribosomal 80s-eef2-sordarin complex from2 yeast obtained by docking atomic models for rna and protein3 components into a 11.7 a cryo-em map. this file, 1s1i,4 contains 60s subunit. the 40s ribosomal subunit is in file5 1s1h.
|
| 43 | c2dycA_ |
|
not modelled |
11.1 |
20 |
PDB header:sugar binding protein
Chain: A: PDB Molecule:galectin-4;
PDBTitle: crystal structure of the n-terminal domain of mouse galectin-4
|
| 44 | d1ulea_ |
|
not modelled |
10.9 |
12 |
Fold:Concanavalin A-like lectins/glucanases
Superfamily:Concanavalin A-like lectins/glucanases
Family:Galectin (animal S-lectin) |
| 45 | d2f0ca1 |
|
not modelled |
10.6 |
20 |
Fold:Virus attachment protein globular domain
Superfamily:Virus attachment protein globular domain
Family:Lactophage receptor-binding protein head domain |
| 46 | c3d8mA_ |
|
not modelled |
10.4 |
24 |
PDB header:virus/viral protein
Chain: A: PDB Molecule:baseplate protein, receptor binding protein;
PDBTitle: crystal structure of a chimeric receptor binding protein from2 lactococcal phages subspecies tp901-1 and p2
|
| 47 | d1s04a_ |
|
not modelled |
10.2 |
22 |
Fold:PUA domain-like
Superfamily:PUA domain-like
Family:ProFAR isomerase associated domain |
| 48 | c3iz5U_ |
|
not modelled |
9.4 |
29 |
PDB header:ribosome
Chain: U: PDB Molecule:60s ribosomal protein l21 (l21e);
PDBTitle: localization of the large subunit ribosomal proteins into a 5.5 a2 cryo-em map of triticum aestivum translating 80s ribosome
|
| 49 | d1r9fa_ |
|
not modelled |
9.4 |
36 |
Fold:Tombusvirus P19 core protein, VP19
Superfamily:Tombusvirus P19 core protein, VP19
Family:Tombusvirus P19 core protein, VP19 |
| 50 | c2p6yA_ |
|
not modelled |
9.3 |
22 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:hypothetical protein vca0587;
PDBTitle: x-ray structure of the protein q9km02_vibch from vibrio cholerae at2 the resolution 1.63 a. northeast structural genomics consortium3 target vcr80.
|
| 51 | c2r7fA_ |
|
not modelled |
9.3 |
5 |
PDB header:hydrolase
Chain: A: PDB Molecule:ribonuclease ii family protein;
PDBTitle: crystal structure of ribonuclease ii family protein from deinococcus2 radiodurans, hexagonal crystal form. northeast structural genomics3 target drr63
|
| 52 | c3izcU_ |
|
not modelled |
9.2 |
35 |
PDB header:ribosome
Chain: U: PDB Molecule:60s ribosomal protein rpl21 (l21e);
PDBTitle: localization of the large subunit ribosomal proteins into a 6.1 a2 cryo-em map of saccharomyces cerevisiae translating 80s ribosome
|
| 53 | c4a1aP_ |
|
not modelled |
9.0 |
35 |
PDB header:ribosome
Chain: P: PDB Molecule:60s ribosomal protein l21;
PDBTitle: t.thermophila 60s ribosomal subunit in complex with2 initiation factor 6. this file contains 5s rrna,3 5.8s rrna and proteins of molecule 3.
|
| 54 | d1cb8a2 |
|
not modelled |
8.8 |
13 |
Fold:Hyaluronate lyase-like, C-terminal domain
Superfamily:Hyaluronate lyase-like, C-terminal domain
Family:Hyaluronate lyase-like, C-terminal domain |
| 55 | d1fnda1 |
|
not modelled |
8.7 |
17 |
Fold:Reductase/isomerase/elongation factor common domain
Superfamily:Riboflavin synthase domain-like
Family:Ferredoxin reductase FAD-binding domain-like |
| 56 | c2wg6L_ |
|
not modelled |
8.1 |
22 |
PDB header:transcription,hydrolase
Chain: L: PDB Molecule:general control protein gcn4,
PDBTitle: proteasome-activating nucleotidase (pan) n-domain (57-134)2 from archaeoglobus fulgidus fused to gcn4, p61a mutant
|
| 57 | c3htnA_ |
|
not modelled |
7.9 |
17 |
PDB header:metal binding protein
Chain: A: PDB Molecule:putative dna binding protein;
PDBTitle: crystal structure of a putative dna binding protein (bt_1116) from2 bacteroides thetaiotaomicron vpi-5482 at 1.50 a resolution
|
| 58 | d1krha1 |
|
not modelled |
7.8 |
16 |
Fold:Reductase/isomerase/elongation factor common domain
Superfamily:Riboflavin synthase domain-like
Family:Ferredoxin reductase FAD-binding domain-like |
| 59 | c2z14A_ |
|
not modelled |
7.7 |
13 |
PDB header:signaling protein
Chain: A: PDB Molecule:ef-hand domain-containing family member c2;
PDBTitle: crystal structure of the n-terminal duf1126 in human ef-2 hand domain containing 2 protein
|
| 60 | d1gawa1 |
|
not modelled |
7.6 |
17 |
Fold:Reductase/isomerase/elongation factor common domain
Superfamily:Riboflavin synthase domain-like
Family:Ferredoxin reductase FAD-binding domain-like |
| 61 | c3ca9A_ |
|
not modelled |
7.5 |
21 |
PDB header:hydrolase
Chain: A: PDB Molecule:deoxyuridine triphosphatase;
PDBTitle: evolution of chlorella virus dutpase
|
| 62 | d2plia1 |
|
not modelled |
7.5 |
21 |
Fold:FAD-binding/transporter-associated domain-like
Superfamily:FAD-binding/transporter-associated domain-like
Family:CorC/HlyC domain-like |
| 63 | d2c9aa2 |
|
not modelled |
7.4 |
23 |
Fold:Concanavalin A-like lectins/glucanases
Superfamily:Concanavalin A-like lectins/glucanases
Family:MAM domain |
| 64 | d2oaia1 |
|
not modelled |
7.0 |
13 |
Fold:FAD-binding/transporter-associated domain-like
Superfamily:FAD-binding/transporter-associated domain-like
Family:CorC/HlyC domain-like |
| 65 | d1pk6a_ |
|
not modelled |
6.9 |
17 |
Fold:TNF-like
Superfamily:TNF-like
Family:TNF-like |
| 66 | c1aqfB_ |
|
not modelled |
6.9 |
17 |
PDB header:transferase
Chain: B: PDB Molecule:pyruvate kinase;
PDBTitle: pyruvate kinase from rabbit muscle with mg, k, and l-2 phospholactate
|
| 67 | d2p13a1 |
|
not modelled |
6.7 |
14 |
Fold:FAD-binding/transporter-associated domain-like
Superfamily:FAD-binding/transporter-associated domain-like
Family:CorC/HlyC domain-like |
| 68 | c3ftjA_ |
|
not modelled |
6.7 |
11 |
PDB header:hydrolase
Chain: A: PDB Molecule:macrolide export atp-binding/permease protein
PDBTitle: crystal structure of the periplasmic region of macb from2 actinobacillus actinomycetemcomitans
|
| 69 | c3m9bK_ |
|
not modelled |
6.7 |
25 |
PDB header:chaperone
Chain: K: PDB Molecule:proteasome-associated atpase;
PDBTitle: crystal structure of the amino terminal coiled coil domain and the2 inter domain of the mycobacterium tuberculosis proteasomal atpase mpa
|
| 70 | d1k8kg_ |
|
not modelled |
6.5 |
14 |
Fold:alpha-alpha superhelix
Superfamily:Arp2/3 complex 16 kDa subunit ARPC5
Family:Arp2/3 complex 16 kDa subunit ARPC5 |
| 71 | d1fdra1 |
|
not modelled |
6.4 |
16 |
Fold:Reductase/isomerase/elongation factor common domain
Superfamily:Riboflavin synthase domain-like
Family:Ferredoxin reductase FAD-binding domain-like |
| 72 | c1t5aB_ |
|
not modelled |
6.3 |
17 |
PDB header:transferase
Chain: B: PDB Molecule:pyruvate kinase, m2 isozyme;
PDBTitle: human pyruvate kinase m2
|
| 73 | c2khjA_ |
|
not modelled |
6.3 |
13 |
PDB header:ribosomal protein
Chain: A: PDB Molecule:30s ribosomal protein s1;
PDBTitle: nmr structure of the domain 6 of the e. coli ribosomal2 protein s1
|
| 74 | d1j8hd1 |
|
not modelled |
6.0 |
27 |
Fold:Immunoglobulin-like beta-sandwich
Superfamily:Immunoglobulin
Family:V set domains (antibody variable domain-like) |
| 75 | d2plsa1 |
|
not modelled |
6.0 |
17 |
Fold:FAD-binding/transporter-associated domain-like
Superfamily:FAD-binding/transporter-associated domain-like
Family:CorC/HlyC domain-like |
| 76 | c2k4vA_ |
|
not modelled |
5.9 |
27 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:uncharacterized protein pa1076;
PDBTitle: solution structure of uncharacterized protein pa1076 from2 pseudomonas aeruginosa. northeast structural genomics3 consortium (nesg) target pat3, ontario center for4 structural proteomics target pa1076 .
|
| 77 | d1duna_ |
|
not modelled |
5.8 |
13 |
Fold:beta-clip
Superfamily:dUTPase-like
Family:dUTPase-like |
| 78 | c2l0dA_ |
|
not modelled |
5.8 |
12 |
PDB header:cell adhesion
Chain: A: PDB Molecule:cell surface protein;
PDBTitle: solution nmr structure of putative cell surface protein ma_4588 (272-2 376 domain) from methanosarcina acetivorans, northeast structural3 genomics consortium target mvr254a
|
| 79 | d1qfza1 |
|
not modelled |
5.6 |
17 |
Fold:Reductase/isomerase/elongation factor common domain
Superfamily:Riboflavin synthase domain-like
Family:Ferredoxin reductase FAD-binding domain-like |
| 80 | c3lgjA_ |
|
not modelled |
5.4 |
20 |
PDB header:dna binding protein
Chain: A: PDB Molecule:single-stranded dna-binding protein;
PDBTitle: crystal structure of single-stranded binding protein (ssb) from2 bartonella henselae
|
| 81 | c2zzfA_ |
|
not modelled |
5.3 |
18 |
PDB header:ligase
Chain: A: PDB Molecule:alanyl-trna synthetase;
PDBTitle: crystal structure of alanyl-trna synthetase without2 oligomerization domain
|
| 82 | c2aklA_ |
|
not modelled |
5.3 |
30 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:phna-like protein pa0128;
PDBTitle: solution structure for phn-a like protein pa0128 from2 pseudomonas aeruginosa
|
| 83 | d3deda1 |
|
not modelled |
5.1 |
17 |
Fold:FAD-binding/transporter-associated domain-like
Superfamily:FAD-binding/transporter-associated domain-like
Family:CorC/HlyC domain-like |
| 84 | d1am2a_ |
|
not modelled |
5.0 |
19 |
Fold:Hedgehog/intein (Hint) domain
Superfamily:Hedgehog/intein (Hint) domain
Family:Intein (protein splicing domain) |
| 85 | c2vl6C_ |
|
not modelled |
5.0 |
27 |
PDB header:dna binding protein
Chain: C: PDB Molecule:minichromosome maintenance protein mcm;
PDBTitle: structural analysis of the sulfolobus solfataricus mcm2 protein n-terminal domain
|