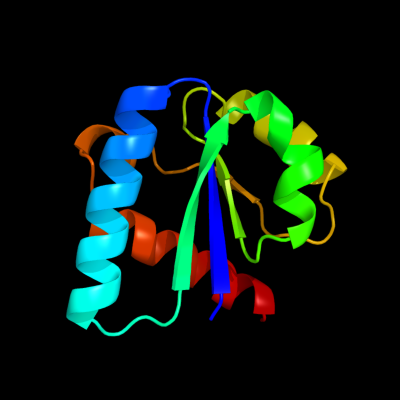
| 1 | d1iiba_
|
|
 |
100.0 |
99 |
Fold:Phosphotyrosine protein phosphatases I-like
Superfamily:PTS system IIB component-like
Family:PTS system, Lactose/Cellobiose specific IIB subunit |
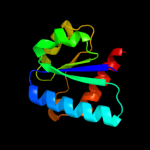
| 2 | c2l2qA_
|
|
 |
100.0 |
35 |
PDB header:transferase
Chain: A: PDB Molecule:pts system, cellobiose-specific iib component (cela);
PDBTitle: solution structure of cellobiose-specific phosphotransferase iib2 component protein from borrelia burgdorferi
|
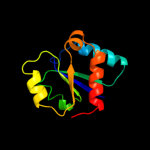
| 3 | c3nbmA_
|
|
 |
100.0 |
14 |
PDB header:transferase
Chain: A: PDB Molecule:pts system, lactose-specific iibc components;
PDBTitle: the lactose-specific iib component domain structure of the2 phosphoenolpyruvate:carbohydrate phosphotransferase system (pts) from3 streptococcus pneumoniae.
|
| 4 | c1tvmA_
|
|
 |
99.3 |
11 |
PDB header:transferase
Chain: A: PDB Molecule:pts system, galactitol-specific iib component;
PDBTitle: nmr structure of enzyme gatb of the galactitol-specific2 phosphoenolpyruvate-dependent phosphotransferase system
|
| 5 | c3czcA_
|
|
 |
99.1 |
16 |
PDB header:transferase
Chain: A: PDB Molecule:rmpb;
PDBTitle: the crystal structure of a putative pts iib(ptxb) from2 streptococcus mutans
|
| 6 | c1vkrA_
|
|
 |
98.7 |
13 |
PDB header:transferase
Chain: A: PDB Molecule:mannitol-specific pts system enzyme iiabc components;
PDBTitle: structure of iib domain of the mannitol-specific permease enzyme ii
|
| 7 | d1vkra_
|
|
 |
98.7 |
13 |
Fold:Phosphotyrosine protein phosphatases I-like
Superfamily:PTS system IIB component-like
Family:PTS system, Lactose/Cellobiose specific IIB subunit |
| 8 | c2kyrA_
|
|
 |
96.5 |
15 |
PDB header:transferase
Chain: A: PDB Molecule:fructose-like phosphotransferase enzyme iib component 1;
PDBTitle: solution structure of enzyme iib subunit of pts system from2 escherichia coli k12. northeast structural genomics consortium target3 er315/ontario center for structural proteomics target ec0544
|
| 9 | d2r48a1
|
|
 |
96.5 |
13 |
Fold:Phosphotyrosine protein phosphatases I-like
Superfamily:PTS system IIB component-like
Family:PTS system, Fructose specific IIB subunit-like |
| 10 | d1vmea1
|
|
 |
96.3 |
17 |
Fold:Flavodoxin-like
Superfamily:Flavoproteins
Family:Flavodoxin-related |
| 11 | c3fniA_
|
|
 |
96.2 |
14 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:putative diflavin flavoprotein a 3;
PDBTitle: crystal structure of a diflavin flavoprotein a3 (all3895) from nostoc2 sp., northeast structural genomics consortium target nsr431a
|
| 12 | d1ycga1
|
|
 |
96.0 |
21 |
Fold:Flavodoxin-like
Superfamily:Flavoproteins
Family:Flavodoxin-related |
| 13 | d2r4qa1
|
|
 |
95.7 |
14 |
Fold:Phosphotyrosine protein phosphatases I-like
Superfamily:PTS system IIB component-like
Family:PTS system, Fructose specific IIB subunit-like |
| 14 | c3sqnB_
|
|
 |
95.7 |
15 |
PDB header:transcription regulator
Chain: B: PDB Molecule:conserved domain protein;
PDBTitle: putative mga family transcriptional regulator from enterococcus2 faecalis
|
| 15 | d1e5da1
|
|
 |
95.3 |
17 |
Fold:Flavodoxin-like
Superfamily:Flavoproteins
Family:Flavodoxin-related |
| 16 | c2ohiB_
|
|
 |
94.9 |
17 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:type a flavoprotein fpra;
PDBTitle: crystal structure of coenzyme f420h2 oxidase (fpra), a diiron2 flavoprotein, reduced state
|
| 17 | c3rofA_
|
|
 |
94.1 |
10 |
PDB header:hydrolase
Chain: A: PDB Molecule:low molecular weight protein-tyrosine-phosphatase ptpa;
PDBTitle: crystal structure of the s. aureus protein tyrosine phosphatase ptpa
|
| 18 | c1vmeB_
|
|
 |
93.8 |
16 |
PDB header:electron transport
Chain: B: PDB Molecule:flavoprotein;
PDBTitle: crystal structure of flavoprotein (tm0755) from thermotoga maritima at2 1.80 a resolution
|
| 19 | c2gi4A_
|
|
 |
93.6 |
18 |
PDB header:hydrolase
Chain: A: PDB Molecule:possible phosphotyrosine protein phosphatase;
PDBTitle: solution structure of the low molecular weight protein2 tyrosine phosphatase from campylobacter jejuni.
|
| 20 | d2arka1
|
|
 |
93.4 |
10 |
Fold:Flavodoxin-like
Superfamily:Flavoproteins
Family:WrbA-like |
| 21 | c3hlyA_ |
|
not modelled |
93.4 |
20 |
PDB header:flavoprotein
Chain: A: PDB Molecule:flavodoxin-like domain;
PDBTitle: crystal structure of the flavodoxin-like domain from2 synechococcus sp q5mzp6_synp6 protein. northeast structural3 genomics consortium target snr135d.
|
| 22 | c3d7nA_ |
|
not modelled |
91.1 |
16 |
PDB header:electron transport
Chain: A: PDB Molecule:flavodoxin, wrba-like protein;
PDBTitle: the crystal structure of the flavodoxin, wrba-like protein from2 agrobacterium tumefaciens
|
| 23 | d2fz5a1 |
|
not modelled |
90.8 |
21 |
Fold:Flavodoxin-like
Superfamily:Flavoproteins
Family:Flavodoxin-related |
| 24 | c1ychD_ |
|
not modelled |
88.8 |
19 |
PDB header:oxidoreductase
Chain: D: PDB Molecule:nitric oxide reductase;
PDBTitle: x-ray crystal structures of moorella thermoacetica fpra.2 novel diiron site structure and mechanistic insights into3 a scavenging nitric oxide reductase
|
| 25 | c1zggA_ |
|
not modelled |
86.7 |
15 |
PDB header:hydrolase
Chain: A: PDB Molecule:putative low molecular weight protein-tyrosine-
PDBTitle: solution structure of a low molecular weight protein2 tyrosine phosphatase from bacillus subtilis
|
| 26 | c3rpeA_ |
|
not modelled |
86.6 |
16 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:modulator of drug activity b;
PDBTitle: 1.1 angstrom crystal structure of putative modulator of drug activity2 (mdab) from yersinia pestis co92.
|
| 27 | d1fmfa_ |
|
not modelled |
86.3 |
17 |
Fold:Flavodoxin-like
Superfamily:Cobalamin (vitamin B12)-binding domain
Family:Cobalamin (vitamin B12)-binding domain |
| 28 | d5pnta_ |
|
not modelled |
85.9 |
10 |
Fold:Phosphotyrosine protein phosphatases I-like
Superfamily:Phosphotyrosine protein phosphatases I
Family:Low-molecular-weight phosphotyrosine protein phosphatases |
| 29 | d1d1qa_ |
|
not modelled |
85.9 |
15 |
Fold:Phosphotyrosine protein phosphatases I-like
Superfamily:Phosphotyrosine protein phosphatases I
Family:Low-molecular-weight phosphotyrosine protein phosphatases |
| 30 | c2l18A_ |
|
not modelled |
85.8 |
8 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:arsenate reductase;
PDBTitle: an arsenate reductase in the phosphate binding state
|
| 31 | d1dg9a_ |
|
not modelled |
85.7 |
13 |
Fold:Phosphotyrosine protein phosphatases I-like
Superfamily:Phosphotyrosine protein phosphatases I
Family:Low-molecular-weight phosphotyrosine protein phosphatases |
| 32 | c3f6sI_ |
|
not modelled |
85.4 |
15 |
PDB header:electron transport
Chain: I: PDB Molecule:flavodoxin;
PDBTitle: desulfovibrio desulfuricans (atcc 29577) oxidized flavodoxin2 alternate conformers
|
| 33 | c3cf4G_ |
|
not modelled |
85.2 |
14 |
PDB header:oxidoreductase
Chain: G: PDB Molecule:acetyl-coa decarboxylase/synthase epsilon subunit;
PDBTitle: structure of the codh component of the m. barkeri acds complex
|
| 34 | d1p3da1 |
|
not modelled |
85.1 |
20 |
Fold:MurCD N-terminal domain
Superfamily:MurCD N-terminal domain
Family:MurCD N-terminal domain |
| 35 | c2hnbA_ |
|
not modelled |
84.9 |
9 |
PDB header:electron transport
Chain: A: PDB Molecule:protein mioc;
PDBTitle: solution structure of a bacterial holo-flavodoxin
|
| 36 | d1f4pa_ |
|
not modelled |
84.0 |
17 |
Fold:Flavodoxin-like
Superfamily:Flavoproteins
Family:Flavodoxin-related |
| 37 | d5nula_ |
|
not modelled |
82.8 |
15 |
Fold:Flavodoxin-like
Superfamily:Flavoproteins
Family:Flavodoxin-related |
| 38 | c2f00A_ |
|
not modelled |
82.5 |
18 |
PDB header:ligase
Chain: A: PDB Molecule:udp-n-acetylmuramate--l-alanine ligase;
PDBTitle: escherichia coli murc
|
| 39 | d1ydga_ |
|
not modelled |
81.7 |
13 |
Fold:Flavodoxin-like
Superfamily:Flavoproteins
Family:WrbA-like |
| 40 | c1u2pA_ |
|
not modelled |
80.0 |
16 |
PDB header:hydrolase
Chain: A: PDB Molecule:low molecular weight protein-tyrosine-
PDBTitle: crystal structure of mycobacterium tuberculosis low2 molecular protein tyrosine phosphatase (mptpa) at 1.9a3 resolution
|
| 41 | c3jviA_ |
|
not modelled |
79.9 |
11 |
PDB header:hydrolase
Chain: A: PDB Molecule:protein tyrosine phosphatase;
PDBTitle: product state mimic crystal structure of protein tyrosine phosphatase2 from entamoeba histolytica
|
| 42 | d1t0ia_ |
|
not modelled |
79.7 |
13 |
Fold:Flavodoxin-like
Superfamily:Flavoproteins
Family:NADPH-dependent FMN reductase |
| 43 | c1gqqA_ |
|
not modelled |
77.9 |
24 |
PDB header:cell wall biosynthesis
Chain: A: PDB Molecule:udp-n-acetylmuramate-l-alanine ligase;
PDBTitle: murc - crystal structure of the apo-enzyme from haemophilus2 influenzae
|
| 44 | c2q9uB_ |
|
not modelled |
77.9 |
14 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:a-type flavoprotein;
PDBTitle: crystal structure of the flavodiiron protein from giardia2 intestinalis
|
| 45 | c2zkiH_ |
|
not modelled |
77.1 |
30 |
PDB header:transcription
Chain: H: PDB Molecule:199aa long hypothetical trp repressor binding
PDBTitle: crystal structure of hypothetical trp repressor binding2 protein from sul folobus tokodaii (st0872)
|
| 46 | d1qrda_ |
|
not modelled |
77.0 |
20 |
Fold:Flavodoxin-like
Superfamily:Flavoproteins
Family:Quinone reductase |
| 47 | c2cwdA_ |
|
not modelled |
76.3 |
8 |
PDB header:hydrolase
Chain: A: PDB Molecule:low molecular weight phosphotyrosine protein phosphatase;
PDBTitle: crystal structure of tt1001 protein from thermus thermophilus hb8
|
| 48 | d1t5ba_ |
|
not modelled |
76.2 |
22 |
Fold:Flavodoxin-like
Superfamily:Flavoproteins
Family:Quinone reductase |
| 49 | d2qwxa1 |
|
not modelled |
74.5 |
19 |
Fold:Flavodoxin-like
Superfamily:Flavoproteins
Family:Quinone reductase |
| 50 | c2g76A_ |
|
not modelled |
73.9 |
13 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:d-3-phosphoglycerate dehydrogenase;
PDBTitle: crystal structure of human 3-phosphoglycerate dehydrogenase
|
| 51 | d1q6za1 |
|
not modelled |
73.5 |
25 |
Fold:DHS-like NAD/FAD-binding domain
Superfamily:DHS-like NAD/FAD-binding domain
Family:Pyruvate oxidase and decarboxylase, middle domain |
| 52 | d2a5la1 |
|
not modelled |
73.3 |
15 |
Fold:Flavodoxin-like
Superfamily:Flavoproteins
Family:WrbA-like |
| 53 | c3edoA_ |
|
not modelled |
73.1 |
14 |
PDB header:flavoprotein
Chain: A: PDB Molecule:putative trp repressor binding protein;
PDBTitle: crystal structure of flavoprotein in complex with fmn2 (yp_193882.1) from lactobacillus acidophilus ncfm at 1.203 a resolution
|
| 54 | c3pg8B_ |
|
not modelled |
72.9 |
23 |
PDB header:transferase
Chain: B: PDB Molecule:phospho-2-dehydro-3-deoxyheptonate aldolase;
PDBTitle: truncated form of 3-deoxy-d-arabino-heptulosonate 7-phosphate synthase2 from thermotoga maritima
|
| 55 | d1ccwa_ |
|
not modelled |
70.5 |
19 |
Fold:Flavodoxin-like
Superfamily:Cobalamin (vitamin B12)-binding domain
Family:Cobalamin (vitamin B12)-binding domain |
| 56 | d1q1aa_ |
|
not modelled |
70.3 |
36 |
Fold:DHS-like NAD/FAD-binding domain
Superfamily:DHS-like NAD/FAD-binding domain
Family:Sir2 family of transcriptional regulators |
| 57 | d1s5pa_ |
|
not modelled |
69.0 |
27 |
Fold:DHS-like NAD/FAD-binding domain
Superfamily:DHS-like NAD/FAD-binding domain
Family:Sir2 family of transcriptional regulators |
| 58 | c3lcmB_ |
|
not modelled |
68.9 |
17 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:putative oxidoreductase;
PDBTitle: crystal structure of smu.1420 from streptococcus mutans ua159
|
| 59 | c2jrlA_ |
|
not modelled |
67.9 |
20 |
PDB header:transcription
Chain: A: PDB Molecule:transcriptional regulator (ntrc family);
PDBTitle: solution structure of the beryllofluoride-activated ntrc4 receiver2 domain dimer
|
| 60 | c2fekA_ |
|
not modelled |
66.5 |
14 |
PDB header:hydrolase
Chain: A: PDB Molecule:low molecular weight protein-tyrosine-
PDBTitle: structure of a protein tyrosine phosphatase
|
| 61 | c1e5dA_ |
|
not modelled |
66.0 |
17 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:rubredoxin\:oxygen oxidoreductase;
PDBTitle: rubredoxin oxygen:oxidoreductase (roo) from anaerobe2 desulfovibrio gigas
|
| 62 | c3ia7A_ |
|
not modelled |
65.9 |
16 |
PDB header:transferase
Chain: A: PDB Molecule:calg4;
PDBTitle: crystal structure of calg4, the calicheamicin glycosyltransferase
|
| 63 | d1jf8a_ |
|
not modelled |
64.1 |
14 |
Fold:Phosphotyrosine protein phosphatases I-like
Superfamily:Phosphotyrosine protein phosphatases I
Family:Low-molecular-weight phosphotyrosine protein phosphatases |
| 64 | c3e18A_ |
|
not modelled |
63.1 |
17 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:oxidoreductase;
PDBTitle: crystal structure of nad-binding protein from listeria innocua
|
| 65 | c1gpjA_ |
|
not modelled |
61.9 |
17 |
PDB header:reductase
Chain: A: PDB Molecule:glutamyl-trna reductase;
PDBTitle: glutamyl-trna reductase from methanopyrus kandleri
|
| 66 | d2b4ya1 |
|
not modelled |
61.8 |
43 |
Fold:DHS-like NAD/FAD-binding domain
Superfamily:DHS-like NAD/FAD-binding domain
Family:Sir2 family of transcriptional regulators |
| 67 | d1vl2a1 |
|
not modelled |
61.4 |
24 |
Fold:Adenine nucleotide alpha hydrolase-like
Superfamily:Adenine nucleotide alpha hydrolases-like
Family:N-type ATP pyrophosphatases |
| 68 | c2wc1A_ |
|
not modelled |
60.7 |
17 |
PDB header:electron transport
Chain: A: PDB Molecule:flavodoxin;
PDBTitle: three-dimensional structure of the nitrogen fixation2 flavodoxin (niff) from rhodobacter capsulatus at 2.2 a
|
| 69 | d1u11a_ |
|
not modelled |
60.0 |
17 |
Fold:Flavodoxin-like
Superfamily:N5-CAIR mutase (phosphoribosylaminoimidazole carboxylase, PurE)
Family:N5-CAIR mutase (phosphoribosylaminoimidazole carboxylase, PurE) |
| 70 | c2qsjB_ |
|
not modelled |
59.6 |
14 |
PDB header:transcription
Chain: B: PDB Molecule:dna-binding response regulator, luxr family;
PDBTitle: crystal structure of a luxr family dna-binding response2 regulator from silicibacter pomeroyi
|
| 71 | c3t4cD_ |
|
not modelled |
59.3 |
14 |
PDB header:transferase
Chain: D: PDB Molecule:2-dehydro-3-deoxyphosphooctonate aldolase 1;
PDBTitle: crystal structure of 2-dehydro-3-deoxyphosphooctonate aldolase from2 burkholderia ambifaria
|
| 72 | d1hdoa_ |
|
not modelled |
59.1 |
19 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Tyrosine-dependent oxidoreductases |
| 73 | d1pvda1 |
|
not modelled |
59.0 |
17 |
Fold:DHS-like NAD/FAD-binding domain
Superfamily:DHS-like NAD/FAD-binding domain
Family:Pyruvate oxidase and decarboxylase, middle domain |
| 74 | c3glsC_ |
|
not modelled |
58.7 |
29 |
PDB header:hydrolase
Chain: C: PDB Molecule:nad-dependent deacetylase sirtuin-3,
PDBTitle: crystal structure of human sirt3
|
| 75 | c3k35D_ |
|
not modelled |
58.5 |
29 |
PDB header:hydrolase
Chain: D: PDB Molecule:nad-dependent deacetylase sirtuin-6;
PDBTitle: crystal structure of human sirt6
|
| 76 | c2an1D_ |
|
not modelled |
58.1 |
23 |
PDB header:transferase
Chain: D: PDB Molecule:putative kinase;
PDBTitle: structural genomics, the crystal structure of a putative kinase from2 salmonella typhimurim lt2
|
| 77 | c2fw9A_ |
|
not modelled |
58.1 |
17 |
PDB header:lyase
Chain: A: PDB Molecule:n5-carboxyaminoimidazole ribonucleotide mutase;
PDBTitle: structure of pure (n5-carboxyaminoimidazole ribonucleotide mutase)2 h59f from the acidophilic bacterium acetobacter aceti, at ph 8
|
| 78 | d1j8fa_ |
|
not modelled |
58.1 |
36 |
Fold:DHS-like NAD/FAD-binding domain
Superfamily:DHS-like NAD/FAD-binding domain
Family:Sir2 family of transcriptional regulators |
| 79 | c3orsD_ |
|
not modelled |
57.7 |
15 |
PDB header:isomerase,biosynthetic protein
Chain: D: PDB Molecule:n5-carboxyaminoimidazole ribonucleotide mutase;
PDBTitle: crystal structure of n5-carboxyaminoimidazole ribonucleotide mutase2 from staphylococcus aureus
|
| 80 | d1t9ba1 |
|
not modelled |
57.4 |
23 |
Fold:DHS-like NAD/FAD-binding domain
Superfamily:DHS-like NAD/FAD-binding domain
Family:Pyruvate oxidase and decarboxylase, middle domain |
| 81 | c3eqzB_ |
|
not modelled |
57.2 |
14 |
PDB header:structural genomics, unknown function
Chain: B: PDB Molecule:response regulator;
PDBTitle: crystal structure of a response regulator from colwellia2 psychrerythraea
|
| 82 | d1o4va_ |
|
not modelled |
57.1 |
16 |
Fold:Flavodoxin-like
Superfamily:N5-CAIR mutase (phosphoribosylaminoimidazole carboxylase, PurE)
Family:N5-CAIR mutase (phosphoribosylaminoimidazole carboxylase, PurE) |
| 83 | d1m2ka_ |
|
not modelled |
57.1 |
29 |
Fold:DHS-like NAD/FAD-binding domain
Superfamily:DHS-like NAD/FAD-binding domain
Family:Sir2 family of transcriptional regulators |
| 84 | d1rlia_ |
|
not modelled |
57.0 |
15 |
Fold:Flavodoxin-like
Superfamily:Flavoproteins
Family:Hypothetical protein YwqN |
| 85 | c1k97A_ |
|
not modelled |
56.7 |
27 |
PDB header:ligase
Chain: A: PDB Molecule:argininosuccinate synthase;
PDBTitle: crystal structure of e. coli argininosuccinate synthetase in complex2 with aspartate and citrulline
|
| 86 | d1qcza_ |
|
not modelled |
56.3 |
17 |
Fold:Flavodoxin-like
Superfamily:N5-CAIR mutase (phosphoribosylaminoimidazole carboxylase, PurE)
Family:N5-CAIR mutase (phosphoribosylaminoimidazole carboxylase, PurE) |
| 87 | c2yxbA_ |
|
not modelled |
56.1 |
8 |
PDB header:isomerase
Chain: A: PDB Molecule:coenzyme b12-dependent mutase;
PDBTitle: crystal structure of the methylmalonyl-coa mutase alpha-subunit from2 aeropyrum pernix
|
| 88 | d1iowa1 |
|
not modelled |
56.0 |
15 |
Fold:PreATP-grasp domain
Superfamily:PreATP-grasp domain
Family:D-Alanine ligase N-terminal domain |
| 89 | c3sz8D_ |
|
not modelled |
55.9 |
23 |
PDB header:transferase
Chain: D: PDB Molecule:2-dehydro-3-deoxyphosphooctonate aldolase 2;
PDBTitle: crystal structure of 2-dehydro-3-deoxyphosphooctonate aldolase from2 burkholderia pseudomallei
|
| 90 | d1gpja2 |
|
not modelled |
55.7 |
18 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Aminoacid dehydrogenase-like, C-terminal domain |
| 91 | c3lp6D_ |
|
not modelled |
55.5 |
20 |
PDB header:lyase
Chain: D: PDB Molecule:phosphoribosylaminoimidazole carboxylase catalytic subunit;
PDBTitle: crystal structure of rv3275c-e60a from mycobacterium tuberculosis at2 1.7a resolution
|
| 92 | c3zquA_ |
|
not modelled |
55.4 |
17 |
PDB header:lyase
Chain: A: PDB Molecule:probable aromatic acid decarboxylase;
PDBTitle: structure of a probable aromatic acid decarboxylase
|
| 93 | c3i2vA_ |
|
not modelled |
54.0 |
20 |
PDB header:transferase
Chain: A: PDB Molecule:adenylyltransferase and sulfurtransferase mocs3;
PDBTitle: crystal structure of human mocs3 rhodanese-like domain
|
| 94 | d2vvpa1 |
|
not modelled |
53.8 |
11 |
Fold:Ribose/Galactose isomerase RpiB/AlsB
Superfamily:Ribose/Galactose isomerase RpiB/AlsB
Family:Ribose/Galactose isomerase RpiB/AlsB |
| 95 | c3fs2A_ |
|
not modelled |
53.5 |
14 |
PDB header:transferase
Chain: A: PDB Molecule:2-dehydro-3-deoxyphosphooctonate aldolase;
PDBTitle: crystal structure of 2-dehydro-3-deoxyphosphooctonate2 aldolase from bruciella melitensis at 1.85a resolution
|
| 96 | d1b1ca_ |
|
not modelled |
53.3 |
16 |
Fold:Flavodoxin-like
Superfamily:Flavoproteins
Family:Cytochrome p450 reductase N-terminal domain-like |
| 97 | c2ywxA_ |
|
not modelled |
53.2 |
19 |
PDB header:lyase
Chain: A: PDB Molecule:phosphoribosylaminoimidazole carboxylase catalytic subunit;
PDBTitle: crystal structure of phosphoribosylaminoimidazole carboxylase2 catalytic subunit from methanocaldococcus jannaschii
|
| 98 | d2djia1 |
|
not modelled |
52.8 |
31 |
Fold:DHS-like NAD/FAD-binding domain
Superfamily:DHS-like NAD/FAD-binding domain
Family:Pyruvate oxidase and decarboxylase, middle domain |
| 99 | c3r74B_ |
|
not modelled |
52.8 |
19 |
PDB header:lyase, biosynthetic protein
Chain: B: PDB Molecule:anthranilate/para-aminobenzoate synthases component i;
PDBTitle: crystal structure of 2-amino-2-desoxyisochorismate synthase (adic)2 synthase phze from burkholderia lata 383
|
| 100 | d1tlla2 |
|
not modelled |
52.5 |
9 |
Fold:Flavodoxin-like
Superfamily:Flavoproteins
Family:Cytochrome p450 reductase N-terminal domain-like |
| 101 | c3k7pA_ |
|
not modelled |
52.3 |
22 |
PDB header:isomerase
Chain: A: PDB Molecule:ribose 5-phosphate isomerase;
PDBTitle: structure of mutant of ribose 5-phosphate isomerase type b from2 trypanosoma cruzi.
|
| 102 | c3m1pA_ |
|
not modelled |
52.3 |
22 |
PDB header:isomerase
Chain: A: PDB Molecule:ribose 5-phosphate isomerase;
PDBTitle: structure of ribose 5-phosphate isomerase type b from trypanosoma2 cruzi, soaked with allose-6-phosphate
|
| 103 | c1wv9B_ |
|
not modelled |
51.9 |
18 |
PDB header:structural genomics, unknown function
Chain: B: PDB Molecule:rhodanese homolog tt1651;
PDBTitle: crystal structure of rhodanese homolog tt1651 from an2 extremely thermophilic bacterium thermus thermophilus hb8
|
| 104 | c3lvuB_ |
|
not modelled |
51.6 |
10 |
PDB header:transport protein
Chain: B: PDB Molecule:abc transporter, periplasmic substrate-binding protein;
PDBTitle: crystal structure of abc transporter, periplasmic substrate-binding2 protein spo2066 from silicibacter pomeroyi
|
| 105 | c2gcgB_ |
|
not modelled |
51.5 |
13 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:glyoxylate reductase/hydroxypyruvate reductase;
PDBTitle: ternary crystal structure of human glyoxylate2 reductase/hydroxypyruvate reductase
|
| 106 | d2ez9a1 |
|
not modelled |
51.2 |
25 |
Fold:DHS-like NAD/FAD-binding domain
Superfamily:DHS-like NAD/FAD-binding domain
Family:Pyruvate oxidase and decarboxylase, middle domain |
| 107 | c3s5pA_ |
|
not modelled |
50.7 |
33 |
PDB header:isomerase
Chain: A: PDB Molecule:ribose 5-phosphate isomerase;
PDBTitle: crystal structure of ribose-5-phosphate isomerase b rpib from giardia2 lamblia
|
| 108 | c3he8A_ |
|
not modelled |
50.3 |
22 |
PDB header:isomerase
Chain: A: PDB Molecule:ribose-5-phosphate isomerase;
PDBTitle: structural study of clostridium thermocellum ribose-5-phosphate2 isomerase b
|
| 109 | c1y80A_ |
|
not modelled |
49.6 |
15 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:predicted cobalamin binding protein;
PDBTitle: structure of a corrinoid (factor iiim)-binding protein from2 moorella thermoacetica
|
| 110 | c2axqA_ |
|
not modelled |
49.5 |
12 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:saccharopine dehydrogenase;
PDBTitle: apo histidine-tagged saccharopine dehydrogenase (l-glu2 forming) from saccharomyces cerevisiae
|
| 111 | c3a2kB_ |
|
not modelled |
49.3 |
14 |
PDB header:ligase/rna
Chain: B: PDB Molecule:trna(ile)-lysidine synthase;
PDBTitle: crystal structure of tils complexed with trna
|
| 112 | c3dzcA_ |
|
not modelled |
49.2 |
3 |
PDB header:isomerase
Chain: A: PDB Molecule:udp-n-acetylglucosamine 2-epimerase;
PDBTitle: 2.35 angstrom resolution structure of wecb (vc0917), a udp-n-2 acetylglucosamine 2-epimerase from vibrio cholerae.
|
| 113 | c3pkiF_ |
|
not modelled |
49.1 |
29 |
PDB header:hydrolase
Chain: F: PDB Molecule:nad-dependent deacetylase sirtuin-6;
PDBTitle: human sirt6 crystal structure in complex with adp ribose
|
| 114 | c3g5jA_ |
|
not modelled |
49.1 |
9 |
PDB header:nucleotide binding protein
Chain: A: PDB Molecule:putative atp/gtp binding protein;
PDBTitle: crystal structure of n-terminal domain of putative atp/gtp binding2 protein from clostridium difficile 630
|
| 115 | c2ejsA_ |
|
not modelled |
49.1 |
17 |
PDB header:ligase
Chain: A: PDB Molecule:autocrine motility factor receptor, isoform 2;
PDBTitle: solution structure of ruh-076, a human cue domain
|
| 116 | c2pgnA_ |
|
not modelled |
49.0 |
32 |
PDB header:hydrolase
Chain: A: PDB Molecule:cyclohexane-1,2-dione hydrolase (cdh);
PDBTitle: the crystal structure of fad and thdp-dependent cyclohexane-1,2-dione2 hydrolase in complex with cyclohexane-1,2-dione
|
| 117 | d1g8aa_ |
|
not modelled |
49.0 |
15 |
Fold:S-adenosyl-L-methionine-dependent methyltransferases
Superfamily:S-adenosyl-L-methionine-dependent methyltransferases
Family:Fibrillarin homologue |
| 118 | c3b2nA_ |
|
not modelled |
48.8 |
17 |
PDB header:transcription
Chain: A: PDB Molecule:uncharacterized protein q99uf4;
PDBTitle: crystal structure of dna-binding response regulator, luxr family, from2 staphylococcus aureus
|
| 119 | c3trhI_ |
|
not modelled |
48.5 |
12 |
PDB header:lyase
Chain: I: PDB Molecule:phosphoribosylaminoimidazole carboxylase
PDBTitle: structure of a phosphoribosylaminoimidazole carboxylase catalytic2 subunit (pure) from coxiella burnetii
|
| 120 | c2ejbA_ |
|
not modelled |
48.5 |
18 |
PDB header:lyase
Chain: A: PDB Molecule:probable aromatic acid decarboxylase;
PDBTitle: crystal structure of phenylacrylic acid decarboxylase from2 aquifex aeolicus
|