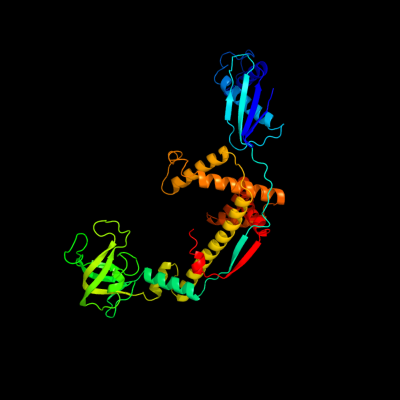
1 c1w26B_
100.0
97
PDB header: chaperoneChain: B: PDB Molecule: trigger factor;PDBTitle: trigger factor in complex with the ribosome forms a2 molecular cradle for nascent proteins
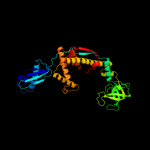
2 c1t11A_
100.0
71
PDB header: chaperoneChain: A: PDB Molecule: trigger factor;PDBTitle: trigger factor
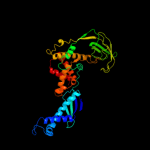
3 c3gtyX_
100.0
20
PDB header: chaperone/ribosomal proteinChain: X: PDB Molecule: trigger factor;PDBTitle: promiscuous substrate recognition in folding and assembly activities2 of the trigger factor chaperone
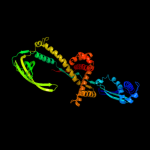
4 d1w26a1
100.0
97
Fold: Triger factor/SurA peptide-binding domain-likeSuperfamily: Triger factor/SurA peptide-binding domain-likeFamily: TF C-terminus5 d1t11a2
100.0
64
Fold: Ribosome binding domain-likeSuperfamily: Trigger factor ribosome-binding domainFamily: Trigger factor ribosome-binding domain6 d1w26a2
100.0
98
Fold: Ribosome binding domain-likeSuperfamily: Trigger factor ribosome-binding domainFamily: Trigger factor ribosome-binding domain7 d1p9ya_
99.9
99
Fold: Ribosome binding domain-likeSuperfamily: Trigger factor ribosome-binding domainFamily: Trigger factor ribosome-binding domain8 d1w26a3
99.9
97
Fold: FKBP-likeSuperfamily: FKBP-likeFamily: FKBP immunophilin/proline isomerase9 c2nsaA_
99.9
13
PDB header: chaperoneChain: A: PDB Molecule: trigger factor;PDBTitle: structures of and interactions between domains of trigger factor from2 themotoga maritim
10 d1t11a3
99.9
74
Fold: FKBP-likeSuperfamily: FKBP-likeFamily: FKBP immunophilin/proline isomerase11 c2nscA_
99.8
23
PDB header: chaperoneChain: A: PDB Molecule: trigger factor;PDBTitle: structures of and interactions between domains of trigger factor from2 themotoga maritima
12 c2d3o1_
99.8
23
PDB header: ribosomeChain: 1: PDB Molecule: trigger factor;PDBTitle: structure of ribosome binding domain of the trigger factor2 on the 50s ribosomal subunit from d. radiodurans
13 d1l1pa_
99.8
99
Fold: FKBP-likeSuperfamily: FKBP-likeFamily: FKBP immunophilin/proline isomerase14 d1t11a1
99.8
75
Fold: Triger factor/SurA peptide-binding domain-likeSuperfamily: Triger factor/SurA peptide-binding domain-likeFamily: TF C-terminus15 d1hxva_
99.7
33
Fold: FKBP-likeSuperfamily: FKBP-likeFamily: FKBP immunophilin/proline isomerase16 c1hxvA_
99.7
33
PDB header: chaperoneChain: A: PDB Molecule: trigger factor;PDBTitle: ppiase domain of the mycoplasma genitalium trigger factor
17 c3prdA_
99.3
25
PDB header: chaperone, isomeraseChain: A: PDB Molecule: fkbp-type peptidyl-prolyl cis-trans isomerase;PDBTitle: structural analysis of protein folding by the methanococcus jannaschii2 chaperone fkbp26
18 c3pr9A_
98.9
29
PDB header: chaperoneChain: A: PDB Molecule: fkbp-type peptidyl-prolyl cis-trans isomerase;PDBTitle: structural analysis of protein folding by the methanococcus jannaschii2 chaperone fkbp26
19 c3cgnA_
98.9
35
PDB header: isomeraseChain: A: PDB Molecule: peptidyl-prolyl cis-trans isomerase;PDBTitle: crystal structure of thermophilic slyd
20 c2kfwA_
98.8
27
PDB header: isomeraseChain: A: PDB Molecule: fkbp-type peptidyl-prolyl cis-trans isomerasePDBTitle: solution structure of full-length slyd from e.coli
21 c2k8iA_
not modelled
98.8
27
PDB header: isomeraseChain: A: PDB Molecule: peptidyl-prolyl cis-trans isomerase;PDBTitle: solution structure of e.coli slyd
22 c2pbcD_
not modelled
98.7
28
PDB header: isomeraseChain: D: PDB Molecule: fk506-binding protein 2;PDBTitle: fk506-binding protein 2
23 d1ix5a_
not modelled
98.5
29
Fold: FKBP-likeSuperfamily: FKBP-likeFamily: FKBP immunophilin/proline isomerase24 c2kr7A_
not modelled
98.5
20
PDB header: isomeraseChain: A: PDB Molecule: fkbp-type peptidyl-prolyl cis-trans isomerase slyd;PDBTitle: solution structure of helicobacter pylori slyd
25 d1c9ha_
not modelled
98.3
22
Fold: FKBP-likeSuperfamily: FKBP-likeFamily: FKBP immunophilin/proline isomerase26 c1q6uA_
not modelled
98.3
23
PDB header: isomeraseChain: A: PDB Molecule: fkbp-type peptidyl-prolyl cis-trans isomerase fkpa;PDBTitle: crystal structure of fkpa from escherichia coli
27 c2vcdA_
not modelled
98.3
25
PDB header: isomeraseChain: A: PDB Molecule: outer membrane protein mip;PDBTitle: solution structure of the fkbp-domain of legionella2 pneumophila mip in complex with rapamycin
28 d1fd9a_
not modelled
98.3
25
Fold: FKBP-likeSuperfamily: FKBP-likeFamily: FKBP immunophilin/proline isomerase29 d1q6ha_
not modelled
98.3
23
Fold: FKBP-likeSuperfamily: FKBP-likeFamily: FKBP immunophilin/proline isomerase30 d1kt0a2
not modelled
98.2
24
Fold: FKBP-likeSuperfamily: FKBP-likeFamily: FKBP immunophilin/proline isomerase31 c2d9fA_
not modelled
98.2
16
PDB header: isomeraseChain: A: PDB Molecule: fk506-binding protein 8 variant;PDBTitle: solution structure of ruh-047, an fkbp domain from human2 cdna
32 d1jvwa_
not modelled
98.2
14
Fold: FKBP-likeSuperfamily: FKBP-likeFamily: FKBP immunophilin/proline isomerase33 d2ppna1
not modelled
98.2
22
Fold: FKBP-likeSuperfamily: FKBP-likeFamily: FKBP immunophilin/proline isomerase34 c2vn1A_
not modelled
98.2
20
PDB header: isomeraseChain: A: PDB Molecule: 70 kda peptidylprolyl isomerase;PDBTitle: crystal structure of the fk506-binding domain of plasmodium2 falciparum fkbp35 in complex with fk506
35 d1yata_
not modelled
98.2
28
Fold: FKBP-likeSuperfamily: FKBP-likeFamily: FKBP immunophilin/proline isomerase36 c2lgoA_
not modelled
98.1
23
PDB header: isomeraseChain: A: PDB Molecule: fkbp;PDBTitle: solution nmr structure of a fkbp-type peptidyl-prolyl cis-trans2 isomerase from giardia lamblia, seattle structural genomics center3 for infectious disease target gilaa.00840.a
37 c3jxvA_
not modelled
98.1
17
PDB header: isomeraseChain: A: PDB Molecule: 70 kda peptidyl-prolyl isomerase;PDBTitle: crystal structure of the 3 fkbp domains of wheat fkbp73
38 c2ke0A_
not modelled
98.1
28
PDB header: isomeraseChain: A: PDB Molecule: peptidyl-prolyl cis-trans isomerase;PDBTitle: solution structure of peptidyl-prolyl cis-trans isomerase from2 burkholderia pseudomallei
39 d1q1ca1
not modelled
98.1
23
Fold: FKBP-likeSuperfamily: FKBP-likeFamily: FKBP immunophilin/proline isomerase40 c1q1cA_
not modelled
98.1
14
PDB header: isomeraseChain: A: PDB Molecule: fk506-binding protein 4;PDBTitle: crystal structure of n(1-260) of human fkbp52
41 c1rouA_
not modelled
98.1
23
PDB header: rotamase (isomerase)Chain: A: PDB Molecule: fkbp59-i;PDBTitle: structure of fkbp59-i, the n-terminal domain of a 59 kda2 fk506-binding protein, nmr, 22 structures
42 c2jwxA_
not modelled
98.1
17
PDB header: apoptosis, isomeraseChain: A: PDB Molecule: fk506-binding protein 8 variant;PDBTitle: solution structure of the n-terminal domain of human fkbp382 (fkbp38ntd)
43 d1kt0a3
not modelled
98.1
14
Fold: FKBP-likeSuperfamily: FKBP-likeFamily: FKBP immunophilin/proline isomerase44 c3o5dB_
not modelled
98.0
14
PDB header: isomeraseChain: B: PDB Molecule: peptidyl-prolyl cis-trans isomerase fkbp5;PDBTitle: crystal structure of a fragment of fkbp51 comprising the fk1 and fk22 domains
45 d1kt1a3
not modelled
98.0
14
Fold: FKBP-likeSuperfamily: FKBP-likeFamily: FKBP immunophilin/proline isomerase46 c3o5fA_
not modelled
98.0
23
PDB header: isomeraseChain: A: PDB Molecule: peptidyl-prolyl cis-trans isomerase fkbp5;PDBTitle: fk1 domain of fkbp51, crystal form vii
47 d1kt1a2
not modelled
98.0
19
Fold: FKBP-likeSuperfamily: FKBP-likeFamily: FKBP immunophilin/proline isomerase48 d1r9ha_
not modelled
97.9
18
Fold: FKBP-likeSuperfamily: FKBP-likeFamily: FKBP immunophilin/proline isomerase49 d1q1ca2
not modelled
97.9
14
Fold: FKBP-likeSuperfamily: FKBP-likeFamily: FKBP immunophilin/proline isomerase50 d1pbka_
not modelled
97.9
23
Fold: FKBP-likeSuperfamily: FKBP-likeFamily: FKBP immunophilin/proline isomerase51 c3oe2A_
not modelled
97.9
21
PDB header: isomeraseChain: A: PDB Molecule: peptidyl-prolyl cis-trans isomerase;PDBTitle: 1.6 a crystal structure of peptidyl-prolyl cis-trans isomerase ppiase2 from pseudomonas syringae pv. tomato str. dc3000 (pspto dc3000)
52 c1zxjB_
not modelled
97.8
19
PDB header: structural genomics, unknown functionChain: B: PDB Molecule: hypothetical protein mg377 homolog;PDBTitle: crystal structure of the hypthetical mycoplasma protein,2 mpn555
53 c1qz2B_
not modelled
97.7
9
PDB header: isomerase/chaperoneChain: B: PDB Molecule: fk506-binding protein 4;PDBTitle: crystal structure of fkbp52 c-terminal domain complex with2 the c-terminal peptide meevd of hsp90
54 c1kt0A_
not modelled
97.7
24
PDB header: isomeraseChain: A: PDB Molecule: 51 kda fk506-binding protein;PDBTitle: structure of the large fkbp-like protein, fkbp51, involved in steroid2 receptor complexes
55 d1m5ya1
not modelled
97.7
12
Fold: Triger factor/SurA peptide-binding domain-likeSuperfamily: Triger factor/SurA peptide-binding domain-likeFamily: Porin chaperone SurA, peptide-binding domain56 c3b7xA_
not modelled
97.6
18
PDB header: isomeraseChain: A: PDB Molecule: fk506-binding protein 6;PDBTitle: crystal structure of human fk506-binding protein 6
57 c2f4eB_
not modelled
97.5
14
PDB header: signaling proteinChain: B: PDB Molecule: atfkbp42;PDBTitle: n-terminal domain of fkbp42 from arabidopsis thaliana
58 d1u79a_
not modelled
96.9
19
Fold: FKBP-likeSuperfamily: FKBP-likeFamily: FKBP immunophilin/proline isomerase59 c1m5yB_
not modelled
96.9
10
PDB header: isomerase, cell cycleChain: B: PDB Molecule: survival protein sura;PDBTitle: crystallographic structure of sura, a molecular chaperone2 that facilitates outer membrane porin folding
60 c2if4A_
not modelled
96.3
12
PDB header: signaling proteinChain: A: PDB Molecule: atfkbp42;PDBTitle: crystal structure of a multi-domain immunophilin from2 arabidopsis thaliana
61 c2pv3B_
not modelled
93.5
14
PDB header: isomeraseChain: B: PDB Molecule: chaperone sura;PDBTitle: crystallographic structure of sura fragment lacking the second2 peptidyl-prolyl isomerase domain complexed with peptide nftlkfwdifrk
62 c3nrkA_
not modelled
91.0
14
PDB header: unknown functionChain: A: PDB Molecule: lic12922;PDBTitle: the crystal structure of the leptospiral hypothetical protein lic12922
63 c2p4vA_
not modelled
90.8
14
PDB header: transcriptionChain: A: PDB Molecule: transcription elongation factor greb;PDBTitle: crystal structure of the transcript cleavage factor, greb2 at 2.6a resolution
64 c3htxA_
not modelled
89.4
17
PDB header: transferase/rnaChain: A: PDB Molecule: hen1;PDBTitle: crystal structure of small rna methyltransferase hen1
65 c3rgcB_
not modelled
88.7
7
PDB header: chaperoneChain: B: PDB Molecule: possible periplasmic protein;PDBTitle: the virulence factor peb4 and the periplasmic protein cj1289 are two2 structurally related sura-like chaperones in the human pathogen3 campylobacter jejuni
66 c1grjA_
not modelled
84.0
14
PDB header: transcription regulationChain: A: PDB Molecule: grea protein;PDBTitle: grea transcript cleavage factor from escherichia coli
67 c3bmbB_
not modelled
79.7
17
PDB header: rna binding proteinChain: B: PDB Molecule: regulator of nucleoside diphosphate kinase;PDBTitle: crystal structure of a new rna polymerase interacting2 protein
68 c2etnA_
not modelled
74.3
14
PDB header: transcriptionChain: A: PDB Molecule: anti-cleavage anti-grea transcription factorPDBTitle: crystal structure of thermus aquaticus gfh1
69 d2f23a2
not modelled
72.2
19
Fold: FKBP-likeSuperfamily: FKBP-likeFamily: GreA transcript cleavage factor, C-terminal domain70 c3s6bA_
not modelled
66.4
16
PDB header: hydrolaseChain: A: PDB Molecule: methionine aminopeptidase;PDBTitle: crystal structure of methionine aminopeptidase 1b from plasmodium2 falciparum, pf10_0150
71 d1jmxa1
not modelled
66.1
8
Fold: Cytochrome cSuperfamily: Cytochrome cFamily: Quinohemoprotein amine dehydrogenase A chain, domains 1 and 272 d2etna2
not modelled
65.6
17
Fold: FKBP-likeSuperfamily: FKBP-likeFamily: GreA transcript cleavage factor, C-terminal domain73 c3rfwA_
not modelled
65.3
24
PDB header: chaperoneChain: A: PDB Molecule: cell-binding factor 2;PDBTitle: the virulence factor peb4 and the periplasmic protein cj1289 are two2 structurally-related sura-like chaperones in the human pathogen3 campylobacter jejuni
74 d1pbya1
not modelled
63.3
5
Fold: Cytochrome cSuperfamily: Cytochrome cFamily: Quinohemoprotein amine dehydrogenase A chain, domains 1 and 275 c2pn0D_
not modelled
55.4
18
PDB header: transcriptionChain: D: PDB Molecule: prokaryotic transcription elongation factorPDBTitle: prokaryotic transcription elongation factor grea/greb from2 nitrosomonas europaea
76 c2wwbA_
not modelled
51.2
11
PDB header: ribosomeChain: A: PDB Molecule: protein transport protein sec61 subunit alpha isoform 1;PDBTitle: cryo-em structure of the mammalian sec61 complex bound to the2 actively translating wheat germ 80s ribosome
77 c3j01A_
not modelled
48.9
17
PDB header: ribosome/ribosomal proteinChain: A: PDB Molecule: preprotein translocase secy subunit;PDBTitle: structure of the ribosome-secye complex in the membrane environment
78 c3dinF_
not modelled
43.6
19
PDB header: membrane protein, protein transportChain: F: PDB Molecule: preprotein translocase subunit secy;PDBTitle: crystal structure of the protein-translocation complex formed by the2 secy channel and the seca atpase
79 d1grja2
not modelled
43.3
15
Fold: FKBP-likeSuperfamily: FKBP-likeFamily: GreA transcript cleavage factor, C-terminal domain80 c2wwaA_
not modelled
42.8
13
PDB header: ribosomeChain: A: PDB Molecule: sec sixty-one protein homolog;PDBTitle: cryo-em structure of idle yeast ssh1 complex bound to the2 yeast 80s ribosome
81 c3dl8H_
not modelled
42.6
24
PDB header: protein transportChain: H: PDB Molecule: preprotein translocase subunit secy;PDBTitle: structure of the complex of aquifex aeolicus secyeg and2 bacillus subtilis seca
82 d1b8za_
not modelled
41.9
32
Fold: IHF-like DNA-binding proteinsSuperfamily: IHF-like DNA-binding proteinsFamily: Prokaryotic DNA-bending protein83 c2akiY_
not modelled
41.0
25
PDB header: protein transportChain: Y: PDB Molecule: preprotein translocase secy subunit;PDBTitle: normal mode-based flexible fitted coordinates of a2 translocating secyeg protein-conducting channel into the3 cryo-em map of a secyeg-nascent chain-70s ribosome complex4 from e. coli
84 c3mp7A_
not modelled
35.8
38
PDB header: protein transportChain: A: PDB Molecule: preprotein translocase subunit secy;PDBTitle: lateral opening of a translocon upon entry of protein suggests the2 mechanism of insertion into membranes
85 c2zqpY_
not modelled
33.8
17
PDB header: protein transportChain: Y: PDB Molecule: preprotein translocase secy subunit;PDBTitle: crystal structure of secye translocon from thermus2 thermophilus
86 c2rhfA_
not modelled
31.0
19
PDB header: hydrolaseChain: A: PDB Molecule: dna helicase recq;PDBTitle: d. radiodurans recq hrdc domain 3
87 c1yw7A_
not modelled
30.8
16
PDB header: hydrolaseChain: A: PDB Molecule: methionine aminopeptidase 2;PDBTitle: h-metap2 complexed with a444148
88 c2q8kA_
not modelled
30.2
26
PDB header: transcriptionChain: A: PDB Molecule: proliferation-associated protein 2g4;PDBTitle: the crystal structure of ebp1
89 c3i18A_
not modelled
28.9
27
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: lmo2051 protein;PDBTitle: crystal structure of the pdz domain of the sdrc-like protein2 (lmo2051) from listeria monocytogenes, northeast structural3 genomics consortium target lmr166b
90 c1b6aA_
not modelled
28.4
19
PDB header: angiogenesis inhibitorChain: A: PDB Molecule: methionine aminopeptidase;PDBTitle: human methionine aminopeptidase 2 complexed with tnp-470
91 c2bpbB_
not modelled
27.8
19
PDB header: oxidoreductaseChain: B: PDB Molecule: sulfite\:cytochrome c oxidoreductase subunit b;PDBTitle: sulfite dehydrogenase from starkeya novella
92 c1yx3A_
not modelled
26.9
15
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: hypothetical protein dsrc;PDBTitle: nmr structure of allochromatium vinosum dsrc: northeast2 structural genomics consortium target op4
93 c2g6pA_
not modelled
25.9
19
PDB header: hydrolaseChain: A: PDB Molecule: methionine aminopeptidase 1;PDBTitle: crystal structure of truncated (delta 1-89) human methionine2 aminopeptidase type 1 in complex with pyridyl pyrimidine derivative
94 c2gz5A_
not modelled
25.9
19
PDB header: hydrolaseChain: A: PDB Molecule: methionine aminopeptidase 1;PDBTitle: human type 1 methionine aminopeptidase in complex with ovalicin at 1.12 ang
95 d1hmja_
not modelled
25.8
16
Fold: RPB5-like RNA polymerase subunitSuperfamily: RPB5-like RNA polymerase subunitFamily: RPB596 d1wuda1
not modelled
24.6
17
Fold: SAM domain-likeSuperfamily: HRDC-likeFamily: HRDC domain from helicases97 d1qxya_
not modelled
23.6
19
Fold: Creatinase/aminopeptidaseSuperfamily: Creatinase/aminopeptidaseFamily: Creatinase/aminopeptidase98 c2xzmO_
not modelled
23.0
11
PDB header: ribosomeChain: O: PDB Molecule: rps13e;PDBTitle: crystal structure of the eukaryotic 40s ribosomal2 subunit in complex with initiation factor 1. this file3 contains the 40s subunit and initiation factor for4 molecule 1
99 c2vcbA_
not modelled
22.7
12
PDB header: hydrolaseChain: A: PDB Molecule: alpha-n-acetylglucosaminidase;PDBTitle: family 89 glycoside hydrolase from clostridium perfringens2 in complex with pugnac