| 1 |
|
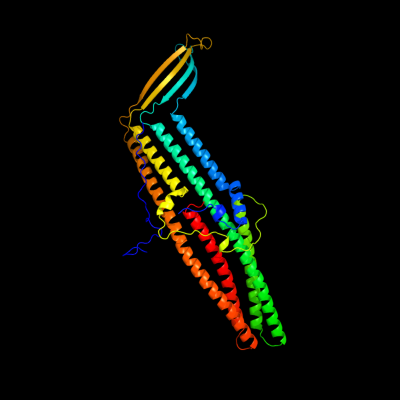
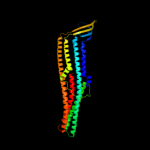
PDB 3pik chain A
Region: 18 - 457
Aligned: 429
Modelled: 440
Confidence: 100.0%
Identity: 100%
PDB header:transport protein
Chain: A: PDB Molecule:cation efflux system protein cusc;
PDBTitle: outer membrane protein cusc
Phyre2
| 2 |
|
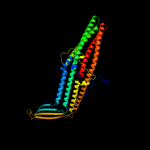
PDB 1wp1 chain A
Region: 18 - 457
Aligned: 440
Modelled: 440
Confidence: 100.0%
Identity: 44%
Fold: Outer membrane efflux proteins (OEP)
Superfamily: Outer membrane efflux proteins (OEP)
Family: Outer membrane efflux proteins (OEP)
Phyre2
| 3 |
|
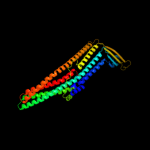
PDB 1yc9 chain A
Region: 49 - 457
Aligned: 403
Modelled: 409
Confidence: 100.0%
Identity: 27%
PDB header:membrane protein
Chain: A: PDB Molecule:multidrug resistance protein;
PDBTitle: the crystal structure of the outer membrane protein vcec from the2 bacterial pathogen vibrio cholerae at 1.8 resolution
Phyre2
| 4 |
|
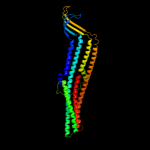
PDB 1tqq chain C
Region: 62 - 455
Aligned: 393
Modelled: 394
Confidence: 100.0%
Identity: 17%
PDB header:transport protein
Chain: C: PDB Molecule:outer membrane protein tolc;
PDBTitle: structure of tolc in complex with hexamminecobalt
Phyre2
| 5 |
|
PDB 1ek9 chain A
Region: 62 - 455
Aligned: 393
Modelled: 391
Confidence: 100.0%
Identity: 17%
Fold: Outer membrane efflux proteins (OEP)
Superfamily: Outer membrane efflux proteins (OEP)
Family: Outer membrane efflux proteins (OEP)
Phyre2
| 6 |
|
PDB 3fpp chain B
Region: 164 - 245
Aligned: 82
Modelled: 82
Confidence: 51.6%
Identity: 15%
PDB header:membrane protein
Chain: B: PDB Molecule:macrolide-specific efflux protein maca;
PDBTitle: crystal structure of e.coli maca
Phyre2
| 7 |
|
PDB 1urq chain C
Region: 180 - 245
Aligned: 66
Modelled: 66
Confidence: 20.8%
Identity: 17%
PDB header:transport protein
Chain: C: PDB Molecule:synaptosomal-associated protein 25;
PDBTitle: crystal structure of neuronal q-snares in complex with2 r-snare motif of tomosyn
Phyre2
| 8 |
|
PDB 3b5n chain C
Region: 185 - 245
Aligned: 61
Modelled: 61
Confidence: 15.4%
Identity: 7%
PDB header:membrane protein
Chain: C: PDB Molecule:protein transport protein sec9;
PDBTitle: structure of the yeast plasma membrane snare complex
Phyre2
| 9 |
|
PDB 3sog chain A
Region: 341 - 455
Aligned: 113
Modelled: 115
Confidence: 10.0%
Identity: 7%
PDB header:structural protein
Chain: A: PDB Molecule:amphiphysin;
PDBTitle: crystal structure of the bar domain of human amphiphysin, isoform 1
Phyre2
| 10 |
|
PDB 1nhl chain A
Region: 194 - 245
Aligned: 52
Modelled: 52
Confidence: 9.8%
Identity: 8%
PDB header:protein transport
Chain: A: PDB Molecule:synaptosomal-associated protein 23;
PDBTitle: snap-23n structure
Phyre2
| 11 |
|
PDB 2f1m chain A
Region: 378 - 453
Aligned: 76
Modelled: 76
Confidence: 8.7%
Identity: 18%
PDB header:transport protein
Chain: A: PDB Molecule:acriflavine resistance protein a;
PDBTitle: conformational flexibility in the multidrug efflux system protein acra
Phyre2
| 12 |
|
PDB 2l06 chain A
Region: 195 - 250
Aligned: 56
Modelled: 56
Confidence: 8.2%
Identity: 16%
PDB header:protein binding
Chain: A: PDB Molecule:phycobilisome lcm core-membrane linker polypeptide;
PDBTitle: solution nmr structure of the pbs linker polypeptide domain (fragment2 254-400) of phycobilisome linker protein apce from synechocystis sp.3 pcc 6803. northeast structural genomics consortium target sgr209c
Phyre2
| 13 |
|
PDB 1l4a chain C
Region: 178 - 242
Aligned: 65
Modelled: 65
Confidence: 7.4%
Identity: 11%
PDB header:endocytosis/exocytosis
Chain: C: PDB Molecule:s-snap25 fusion protein;
PDBTitle: x-ray structure of the neuronal complexin/snare complex2 from the squid loligo pealei
Phyre2
| 14 |
|
PDB 2l3w chain A
Region: 200 - 250
Aligned: 51
Modelled: 51
Confidence: 6.6%
Identity: 20%
PDB header:photosynthesis
Chain: A: PDB Molecule:phycobilisome rod linker polypeptide;
PDBTitle: solution nmr structure of the pbs linker domain of phycobilisome rod2 linker polypeptide from synechococcus elongatus, northeast structural3 genomics consortium target snr168a
Phyre2
| 15 |
|
PDB 3ohw chain B
Region: 192 - 250
Aligned: 59
Modelled: 59
Confidence: 6.6%
Identity: 14%
PDB header:protein binding
Chain: B: PDB Molecule:phycobilisome lcm core-membrane linker polypeptide;
PDBTitle: x-ray structure of phycobilisome lcm core-membrane linker polypeptide2 (fragment 721-860) from synechocystis sp. pcc 6803, northeast3 structural genomics consortium target sgr209e
Phyre2
| 16 |
|
PDB 2v7s chain A
Region: 273 - 359
Aligned: 87
Modelled: 87
Confidence: 6.6%
Identity: 8%
PDB header:unknown function
Chain: A: PDB Molecule:probable conserved lipoprotein lppa;
PDBTitle: crystal structure of the putative lipoprotein lppa from2 mycobacterium tuberculosis
Phyre2
| 17 |
|
PDB 3pru chain D
Region: 200 - 250
Aligned: 51
Modelled: 51
Confidence: 6.4%
Identity: 18%
PDB header:photosynthesis
Chain: D: PDB Molecule:phycobilisome 32.1 kda linker polypeptide, phycocyanin-
PDBTitle: crystal structure of phycobilisome 32.1 kda linker polypeptide,2 phycocyanin-associated, rod 1 (fragment 14-158) from synechocystis3 sp. pcc 6803, northeast structural genomics consortium target sgr182a
Phyre2
| 18 |
|
PDB 2ky4 chain A
Region: 192 - 250
Aligned: 59
Modelled: 59
Confidence: 6.2%
Identity: 14%
PDB header:photosynthesis
Chain: A: PDB Molecule:phycobilisome linker polypeptide;
PDBTitle: solution nmr structure of the pbs linker domain of phycobilisome2 linker polypeptide from anabaena sp. northeast structural genomics3 consortium target nsr123e
Phyre2
| 19 |
|
PDB 1yud chain A domain 1
Region: 15 - 34
Aligned: 20
Modelled: 20
Confidence: 5.9%
Identity: 25%
Fold: Double-stranded beta-helix
Superfamily: RmlC-like cupins
Family: YML079-like
Phyre2
| 20 |
|
PDB 2d7c chain D
Region: 63 - 77
Aligned: 15
Modelled: 15
Confidence: 5.2%
Identity: 13%
PDB header:protein transport
Chain: D: PDB Molecule:rab11 family-interacting protein 3;
PDBTitle: crystal structure of human rab11 in complex with fip3 rab-2 binding domain
Phyre2