1 c2l9uA_
31.7
25
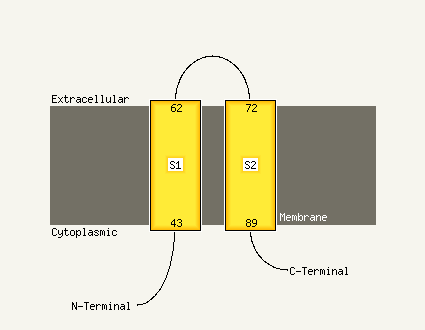
PDB header: membrane proteinChain: A: PDB Molecule: receptor tyrosine-protein kinase erbb-3;PDBTitle: spatial structure of dimeric erbb3 transmembrane domain
2 c3lwfD_
23.2
17
PDB header: transcription regulatorChain: D: PDB Molecule: putative transcriptional regulator;PDBTitle: crystal structure of putative transcriptional regulator (np_470886.1)2 from listeria innocua at 2.06 a resolution
3 c3tr7A_
17.3
35
PDB header: hydrolaseChain: A: PDB Molecule: uracil-dna glycosylase;PDBTitle: structure of a uracil-dna glycosylase (ung) from coxiella burnetii
4 d2osoa1
15.8
21
Fold: Ligand-binding domain in the NO signalling and Golgi transportSuperfamily: Ligand-binding domain in the NO signalling and Golgi transportFamily: MJ1460-like5 c3ajbB_
14.3
31
PDB header: protein transportChain: B: PDB Molecule: peroxisomal biogenesis factor 19;PDBTitle: crystal structure of human pex3p in complex with n-terminal pex19p2 peptide
6 c3njcA_
13.8
31
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: yslb protein;PDBTitle: crystal structure of the yslb protein from bacillus subtilis.2 northeast structural genomics consortium target sr460.
7 c1iojA_
13.2
27
PDB header: apolipoproteinChain: A: PDB Molecule: apoc-i;PDBTitle: human apolipoprotein c-i, nmr, 18 structures
8 d2qtva3
12.9
9
Fold: vWA-likeSuperfamily: vWA-likeFamily: Trunk domain of Sec23/249 c3sibA_
12.3
13
PDB header: dna binding proteinChain: A: PDB Molecule: ure3-bp sequence specific dna binding protein;PDBTitle: crystal structure of ure3-binding protein, wild-type
10 d1tafb_
12.2
20
Fold: Histone-foldSuperfamily: Histone-foldFamily: TBP-associated factors, TAFs11 c1kcfB_
11.6
8
PDB header: hydrolaseChain: B: PDB Molecule: hypothetical 30.2 kd protein c25g10.02 inPDBTitle: crystal structure of the yeast mitochondrial holliday2 junction resolvase, ydc2
12 d3euga_
10.4
32
Fold: Uracil-DNA glycosylase-likeSuperfamily: Uracil-DNA glycosylase-likeFamily: Uracil-DNA glycosylase13 d1okba_
10.0
23
Fold: Uracil-DNA glycosylase-likeSuperfamily: Uracil-DNA glycosylase-likeFamily: Uracil-DNA glycosylase14 d2j8xa1
9.6
23
Fold: Uracil-DNA glycosylase-likeSuperfamily: Uracil-DNA glycosylase-likeFamily: Uracil-DNA glycosylase15 c1p5kA_
9.5
55
PDB header: ribosomeChain: A: PDB Molecule: 19-mer peptide from 50s ribosomal protein l1;PDBTitle: hp (2-20) substitution ser to leu11 modification in sds-d252 micelles
16 d1laue_
9.2
32
Fold: Uracil-DNA glycosylase-likeSuperfamily: Uracil-DNA glycosylase-likeFamily: Uracil-DNA glycosylase17 d1vfga1
9.0
33
Fold: Poly A polymerase C-terminal region-likeSuperfamily: Poly A polymerase C-terminal region-likeFamily: Poly A polymerase C-terminal region-like18 c3ls1A_
8.7
18
PDB header: photosynthesisChain: A: PDB Molecule: sll1638 protein;PDBTitle: crystal structure of cyanobacterial psbq from synechocystis2 sp. pcc 6803 complexed with zn2+
19 c3cxmA_
8.0
27
PDB header: hydrolaseChain: A: PDB Molecule: uracil-dna glycosylase;PDBTitle: leishmania naiffi uracil-dna glycosylase in complex with 5-bromouracil
20 d2hxma1
7.6
23
Fold: Uracil-DNA glycosylase-likeSuperfamily: Uracil-DNA glycosylase-likeFamily: Uracil-DNA glycosylase21 c2booA_
not modelled
7.5
23
PDB header: hydrolaseChain: A: PDB Molecule: uracil-dna glycosylase;PDBTitle: the crystal structure of uracil-dna n-glycosylase (ung)2 from deinococcus radiodurans.
22 d1oeyj_
not modelled
7.2
22
Fold: beta-Grasp (ubiquitin-like)Superfamily: CAD & PB1 domainsFamily: PB1 domain23 d1pd0a3
not modelled
6.3
9
Fold: vWA-likeSuperfamily: vWA-likeFamily: Trunk domain of Sec23/2424 d2gykb1
not modelled
6.3
27
Fold: His-Me finger endonucleasesSuperfamily: His-Me finger endonucleasesFamily: HNH-motif25 c3eh2B_
not modelled
6.1
7
PDB header: protein transportChain: B: PDB Molecule: protein transport protein sec24c;PDBTitle: crystal structure of the human copii-coat protein sec24c
26 c3op5B_
not modelled
6.0
30
PDB header: transferaseChain: B: PDB Molecule: serine/threonine-protein kinase vrk1;PDBTitle: human vaccinia-related kinase 1
27 d1miwa1
not modelled
6.0
17
Fold: Poly A polymerase C-terminal region-likeSuperfamily: Poly A polymerase C-terminal region-likeFamily: Poly A polymerase C-terminal region-like28 c2akhZ_
not modelled
5.9
25
PDB header: protein transportChain: Z: PDB Molecule: preprotein translocase sece subunit;PDBTitle: normal mode-based flexible fitted coordinates of a non-2 translocating secyeg protein-conducting channel into the3 cryo-em map of a secyeg-nascent chain-70s ribosome complex4 from e. coli
29 d1csna_
not modelled
5.7
24
Fold: Protein kinase-like (PK-like)Superfamily: Protein kinase-like (PK-like)Family: Protein kinases, catalytic subunit30 c3dinD_
not modelled
5.6
50
PDB header: membrane protein, protein transportChain: D: PDB Molecule: preprotein translocase subunit sece;PDBTitle: crystal structure of the protein-translocation complex formed by the2 secy channel and the seca atpase
31 d1rbli_
not modelled
5.5
56
Fold: RuBisCO, small subunitSuperfamily: RuBisCO, small subunitFamily: RuBisCO, small subunit32 d1kcfa2
not modelled
5.2
8
Fold: Ribonuclease H-like motifSuperfamily: Ribonuclease H-likeFamily: Mitochondrial resolvase ydc2 catalytic domain