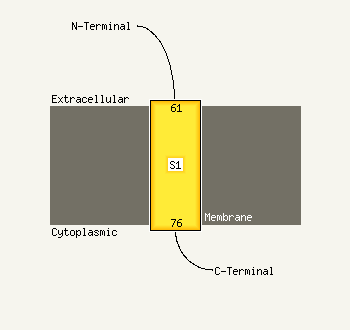
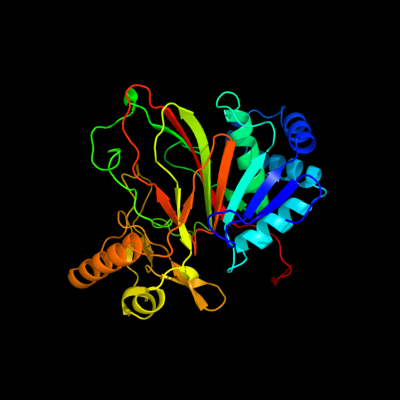
| 1 | d1jr7a_
|
|
 |
100.0 |
99 |
Fold:Double-stranded beta-helix
Superfamily:Clavaminate synthase-like
Family:Gab protein (hypothetical protein YgaT) |
| 2 | c3ms5A_
|
|
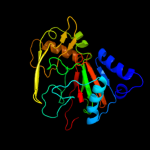 |
100.0 |
17 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:gamma-butyrobetaine dioxygenase;
PDBTitle: crystal structure of human gamma-butyrobetaine,2-oxoglutarate2 dioxygenase 1 (bbox1)
|
| 3 | c2og5A_
|
|
 |
100.0 |
16 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:putative oxygenase;
PDBTitle: crystal structure of asparagine oxygenase (asno)
|
| 4 | d1ds1a_
|
|
 |
100.0 |
17 |
Fold:Double-stranded beta-helix
Superfamily:Clavaminate synthase-like
Family:Clavaminate synthase |
| 5 | c2wbqA_
|
|
 |
100.0 |
19 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:l-arginine beta-hydroxylase;
PDBTitle: crystal structure of vioc in complex with (2s,3s)-2 hydroxyarginine
|
| 6 | d1oiha_
|
|
 |
100.0 |
17 |
Fold:Double-stranded beta-helix
Superfamily:Clavaminate synthase-like
Family:TauD/TfdA-like |
| 7 | d1otja_
|
|
 |
100.0 |
17 |
Fold:Double-stranded beta-helix
Superfamily:Clavaminate synthase-like
Family:TauD/TfdA-like |
| 8 | c3eatX_
|
|
 |
100.0 |
18 |
PDB header:oxidoreductase
Chain: X: PDB Molecule:pyoverdine biosynthesis protein pvcb;
PDBTitle: crystal structure of the pvcb (pa2255) protein from2 pseudomonas aeruginosa
|
| 9 | c3pvjB_
|
|
 |
100.0 |
13 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:alpha-ketoglutarate-dependent taurine dioxygenase;
PDBTitle: crystal structure of the fe(ii)/alpha-ketoglutarate dependent taurine2 dioxygenase from pseudomonas putida kt2440
|
| 10 | c3r1jB_
|
|
 |
100.0 |
16 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:alpha-ketoglutarate-dependent taurine dioxygenase;
PDBTitle: crystal structure of alpha-ketoglutarate-dependent taurine dioxygenase2 from mycobacterium avium, native form
|
| 11 | d1nx4a_
|
|
 |
100.0 |
18 |
Fold:Double-stranded beta-helix
Superfamily:Clavaminate synthase-like
Family:gamma-Butyrobetaine hydroxylase |
| 12 | d1y0za_
|
|
 |
99.9 |
21 |
Fold:Double-stranded beta-helix
Superfamily:Clavaminate synthase-like
Family:gamma-Butyrobetaine hydroxylase |
| 13 | c3pl0B_
|
|
 |
94.1 |
36 |
PDB header:biosynthetic protein
Chain: B: PDB Molecule:uncharacterized protein;
PDBTitle: crystal structure of a bsma homolog (mpe_a2762) from methylobium2 petroleophilum pm1 at 1.91 a resolution
|
| 14 | d1wdia_
|
|
 |
92.5 |
17 |
Fold:QueA-like
Superfamily:QueA-like
Family:QueA-like |
| 15 | c1yy3A_
|
|
 |
92.1 |
33 |
PDB header:isomerase
Chain: A: PDB Molecule:s-adenosylmethionine:trna ribosyltransferase-
PDBTitle: structure of s-adenosylmethionine:trna ribosyltransferase-2 isomerase (quea)
|
| 16 | d1vkya_
|
|
 |
91.0 |
22 |
Fold:QueA-like
Superfamily:QueA-like
Family:QueA-like |
| 17 | d2fcta1
|
|
 |
84.5 |
12 |
Fold:Double-stranded beta-helix
Superfamily:Clavaminate synthase-like
Family:PhyH-like |
| 18 | c2opwA_
|
|
 |
79.5 |
19 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:phyhd1 protein;
PDBTitle: crystal structure of human phytanoyl-coa dioxygenase phyhd1 (apo)
|
| 19 | d2a1xa1
|
|
 |
73.1 |
14 |
Fold:Double-stranded beta-helix
Superfamily:Clavaminate synthase-like
Family:PhyH-like |
| 20 | c2rdsA_
|
|
 |
69.4 |
19 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:1-deoxypentalenic acid 11-beta hydroxylase; fe(ii)/alpha-
PDBTitle: crystal structure of ptlh with fe/oxalylglycine and ent-1-2 deoxypentalenic acid bound
|
| 21 | c3emrA_ |
|
not modelled |
68.5 |
16 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:ectd;
PDBTitle: crystal structure analysis of the ectoine hydroxylase ectd from2 salibacillus salexigens
|
| 22 | c3gjbA_ |
|
not modelled |
67.9 |
13 |
PDB header:biosynthetic protein
Chain: A: PDB Molecule:cytc3;
PDBTitle: cytc3 with fe(ii) and alpha-ketoglutarate
|
| 23 | c3nnlB_ |
|
not modelled |
54.2 |
16 |
PDB header:biosynthetic protein
Chain: B: PDB Molecule:cura;
PDBTitle: halogenase domain from cura module (crystal form iii)
|
| 24 | c2fg0B_ |
|
not modelled |
50.5 |
10 |
PDB header:hydrolase
Chain: B: PDB Molecule:cog0791: cell wall-associated hydrolases (invasion-
PDBTitle: crystal structure of a putative gamma-d-glutamyl-l-diamino acid2 endopeptidase (npun_r0659) from nostoc punctiforme pcc 73102 at 1.793 a resolution
|
| 25 | d1v9la2 |
|
not modelled |
43.8 |
19 |
Fold:Aminoacid dehydrogenase-like, N-terminal domain
Superfamily:Aminoacid dehydrogenase-like, N-terminal domain
Family:Aminoacid dehydrogenases |
| 26 | d2evra2 |
|
not modelled |
37.5 |
9 |
Fold:Cysteine proteinases
Superfamily:Cysteine proteinases
Family:NlpC/P60 |
| 27 | c3npfB_ |
|
not modelled |
32.6 |
19 |
PDB header:hydrolase
Chain: B: PDB Molecule:putative dipeptidyl-peptidase vi;
PDBTitle: crystal structure of a putative dipeptidyl-peptidase vi (bacova_00612)2 from bacteroides ovatus at 1.72 a resolution
|
| 28 | d1dqua_ |
|
not modelled |
29.8 |
25 |
Fold:TIM beta/alpha-barrel
Superfamily:Phosphoenolpyruvate/pyruvate domain
Family:Phosphoenolpyruvate mutase/Isocitrate lyase-like |
| 29 | c2xivA_ |
|
not modelled |
28.6 |
13 |
PDB header:structural protein
Chain: A: PDB Molecule:hypothetical invasion protein;
PDBTitle: structure of rv1477, hypothetical invasion protein of2 mycobacterium tuberculosis
|
| 30 | d1f61a_ |
|
not modelled |
27.7 |
27 |
Fold:TIM beta/alpha-barrel
Superfamily:Phosphoenolpyruvate/pyruvate domain
Family:Phosphoenolpyruvate mutase/Isocitrate lyase-like |
| 31 | d1ywka1 |
|
not modelled |
25.5 |
13 |
Fold:Double-stranded beta-helix
Superfamily:RmlC-like cupins
Family:KduI-like |
| 32 | c3gt2A_ |
|
not modelled |
25.5 |
6 |
PDB header:unknown function
Chain: A: PDB Molecule:putative uncharacterized protein;
PDBTitle: crystal structure of the p60 domain from m. avium2 paratuberculosis antigen map1272c
|
| 33 | c2dbiA_ |
|
not modelled |
23.9 |
11 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:hypothetical protein ybiu;
PDBTitle: crystal structure of a hypothetical protein jw0805 from2 escherichia coli
|
| 34 | c1yo8A_ |
|
not modelled |
23.5 |
13 |
PDB header:cell adhesion
Chain: A: PDB Molecule:thrombospondin-2;
PDBTitle: structure of the c-terminal domain of human thrombospondin-2
|
| 35 | d1xnea_ |
|
not modelled |
19.6 |
17 |
Fold:PUA domain-like
Superfamily:PUA domain-like
Family:ProFAR isomerase associated domain |
| 36 | c3rcqA_ |
|
not modelled |
19.1 |
25 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:aspartyl/asparaginyl beta-hydroxylase;
PDBTitle: crystal structure of human aspartate beta-hydroxylase isoform a
|
| 37 | d2axte1 |
|
not modelled |
18.7 |
36 |
Fold:Single transmembrane helix
Superfamily:Cytochrome b559 subunits
Family:Cytochrome b559 subunits |
| 38 | d1fw8a_ |
|
not modelled |
18.4 |
29 |
Fold:Phosphoglycerate kinase
Superfamily:Phosphoglycerate kinase
Family:Phosphoglycerate kinase |
| 39 | d1s04a_ |
|
not modelled |
18.1 |
6 |
Fold:PUA domain-like
Superfamily:PUA domain-like
Family:ProFAR isomerase associated domain |
| 40 | d16pka_ |
|
not modelled |
18.0 |
21 |
Fold:Phosphoglycerate kinase
Superfamily:Phosphoglycerate kinase
Family:Phosphoglycerate kinase |
| 41 | c3ndhA_ |
|
not modelled |
17.8 |
23 |
PDB header:hydrolase/dna
Chain: A: PDB Molecule:restriction endonuclease thai;
PDBTitle: restriction endonuclease in complex with substrate dna
|
| 42 | c2k1gA_ |
|
not modelled |
17.4 |
20 |
PDB header:lipoprotein
Chain: A: PDB Molecule:lipoprotein spr;
PDBTitle: solution nmr structure of lipoprotein spr from escherichia coli k12.2 northeast structural genomics target er541-37-162
|
| 43 | c3pbiA_ |
|
not modelled |
16.8 |
13 |
PDB header:hydrolase
Chain: A: PDB Molecule:invasion protein;
PDBTitle: structure of the peptidoglycan hydrolase ripb (rv1478) from2 mycobacterium tuberculosis at 1.6 resolution
|
| 44 | c1zmrA_ |
|
not modelled |
16.7 |
21 |
PDB header:transferase
Chain: A: PDB Molecule:phosphoglycerate kinase;
PDBTitle: crystal structure of the e. coli phosphoglycerate kinase
|
| 45 | d1b26a2 |
|
not modelled |
16.7 |
19 |
Fold:Aminoacid dehydrogenase-like, N-terminal domain
Superfamily:Aminoacid dehydrogenase-like, N-terminal domain
Family:Aminoacid dehydrogenases |
| 46 | d1te5a_ |
|
not modelled |
16.6 |
17 |
Fold:Ntn hydrolase-like
Superfamily:N-terminal nucleophile aminohydrolases (Ntn hydrolases)
Family:Class II glutamine amidotransferases |
| 47 | d1l1fa2 |
|
not modelled |
16.2 |
21 |
Fold:Aminoacid dehydrogenase-like, N-terminal domain
Superfamily:Aminoacid dehydrogenase-like, N-terminal domain
Family:Aminoacid dehydrogenases |
| 48 | d1vpea_ |
|
not modelled |
16.1 |
29 |
Fold:Phosphoglycerate kinase
Superfamily:Phosphoglycerate kinase
Family:Phosphoglycerate kinase |
| 49 | d1e5ra_ |
|
not modelled |
15.5 |
6 |
Fold:Double-stranded beta-helix
Superfamily:Clavaminate synthase-like
Family:Type II Proline 3-hydroxylase (proline oxidase) |
| 50 | d1phpa_ |
|
not modelled |
14.9 |
29 |
Fold:Phosphoglycerate kinase
Superfamily:Phosphoglycerate kinase
Family:Phosphoglycerate kinase |
| 51 | d1hdia_ |
|
not modelled |
14.9 |
29 |
Fold:Phosphoglycerate kinase
Superfamily:Phosphoglycerate kinase
Family:Phosphoglycerate kinase |
| 52 | c1ywkE_ |
|
not modelled |
14.7 |
14 |
PDB header:isomerase
Chain: E: PDB Molecule:4-deoxy-l-threo-5-hexosulose-uronate ketol-
PDBTitle: crystal structure of 4-deoxy-1-threo-5-hexosulose-uronate2 ketol-isomerase from enterococcus faecalis
|
| 53 | d1v6sa_ |
|
not modelled |
14.6 |
29 |
Fold:Phosphoglycerate kinase
Superfamily:Phosphoglycerate kinase
Family:Phosphoglycerate kinase |
| 54 | d1qpga_ |
|
not modelled |
14.3 |
29 |
Fold:Phosphoglycerate kinase
Superfamily:Phosphoglycerate kinase
Family:Phosphoglycerate kinase |
| 55 | c3lvwA_ |
|
not modelled |
14.2 |
7 |
PDB header:ligase
Chain: A: PDB Molecule:glutamate--cysteine ligase;
PDBTitle: glutathione-inhibited scgcl
|
| 56 | c3f1zF_ |
|
not modelled |
13.6 |
24 |
PDB header:dna binding protein
Chain: F: PDB Molecule:putative nucleic acid-binding lipoprotein;
PDBTitle: crystal structure of putative nucleic acid-binding lipoprotein2 (yp_001337197.1) from klebsiella pneumoniae subsp. pneumoniae mgh3 78578 at 2.46 a resolution
|
| 57 | d1ltka_ |
|
not modelled |
13.6 |
21 |
Fold:Phosphoglycerate kinase
Superfamily:Phosphoglycerate kinase
Family:Phosphoglycerate kinase |
| 58 | c3uyjA_ |
|
not modelled |
13.6 |
38 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:lysine-specific demethylase 8;
PDBTitle: crystal structure of jmjd5 catalytic core domain in complex with2 nickle and alpha-kg
|
| 59 | c1ux6A_ |
|
not modelled |
13.4 |
11 |
PDB header:cell adhesion
Chain: A: PDB Molecule:thrombospondin-1;
PDBTitle: structure of a thrombospondin c-terminal fragment reveals a2 novel calcium core in the type 3 repeats
|
| 60 | c2jijA_ |
|
not modelled |
13.3 |
24 |
PDB header:hydrolase
Chain: A: PDB Molecule:prolyl-4 hydroxylase;
PDBTitle: crystal structure of the apo form of chlamydomonas2 reinhardtii prolyl-4 hydroxylase type i
|
| 61 | c3i86A_ |
|
not modelled |
12.8 |
15 |
PDB header:hydrolase
Chain: A: PDB Molecule:putative uncharacterized protein;
PDBTitle: crystal structure of the p60 domain from m. avium subspecies2 paratuberculosis antigen map1204
|
| 62 | d1vjda_ |
|
not modelled |
12.8 |
29 |
Fold:Phosphoglycerate kinase
Superfamily:Phosphoglycerate kinase
Family:Phosphoglycerate kinase |
| 63 | c3itqB_ |
|
not modelled |
12.6 |
36 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:prolyl 4-hydroxylase, alpha subunit domain protein;
PDBTitle: crystal structure of a prolyl 4-hydroxylase from bacillus anthracis
|
| 64 | c3aicC_ |
|
not modelled |
11.8 |
23 |
PDB header:transferase
Chain: C: PDB Molecule:glucosyltransferase-si;
PDBTitle: crystal structure of glucansucrase from streptococcus mutans
|
| 65 | c3al6A_ |
|
not modelled |
11.7 |
23 |
PDB header:unknown function
Chain: A: PDB Molecule:jmjc domain-containing protein c2orf60;
PDBTitle: crystal structure of human tyw5
|
| 66 | d2toda1 |
|
not modelled |
11.7 |
33 |
Fold:Domain of alpha and beta subunits of F1 ATP synthase-like
Superfamily:Alanine racemase C-terminal domain-like
Family:Eukaryotic ODC-like |
| 67 | c1wtaA_ |
|
not modelled |
11.6 |
11 |
PDB header:transferase
Chain: A: PDB Molecule:5'-methylthioadenosine phosphorylase;
PDBTitle: crystal structure of 5'-deoxy-5'-methylthioadenosine from aeropyrum2 pernix (r32 form)
|
| 68 | c3q3vA_ |
|
not modelled |
11.5 |
36 |
PDB header:transferase
Chain: A: PDB Molecule:phosphoglycerate kinase;
PDBTitle: crystal structure of phosphoglycerate kinase from campylobacter2 jejuni.
|
| 69 | c2w8iG_ |
|
not modelled |
11.4 |
0 |
PDB header:membrane protein
Chain: G: PDB Molecule:putative outer membrane lipoprotein wza;
PDBTitle: crystal structure of wza24-345.
|
| 70 | c3h41A_ |
|
not modelled |
11.0 |
13 |
PDB header:hydrolase
Chain: A: PDB Molecule:nlp/p60 family protein;
PDBTitle: crystal structure of a nlpc/p60 family protein (bce_2878) from2 bacillus cereus atcc 10987 at 1.79 a resolution
|
| 71 | d1bvua2 |
|
not modelled |
11.0 |
14 |
Fold:Aminoacid dehydrogenase-like, N-terminal domain
Superfamily:Aminoacid dehydrogenase-like, N-terminal domain
Family:Aminoacid dehydrogenases |
| 72 | d1vrba1 |
|
not modelled |
10.9 |
23 |
Fold:Double-stranded beta-helix
Superfamily:Clavaminate synthase-like
Family:Asparaginyl hydroxylase-like |
| 73 | d1njjb1 |
|
not modelled |
10.7 |
36 |
Fold:Domain of alpha and beta subunits of F1 ATP synthase-like
Superfamily:Alanine racemase C-terminal domain-like
Family:Eukaryotic ODC-like |
| 74 | d1cb0a_ |
|
not modelled |
10.5 |
7 |
Fold:Phosphorylase/hydrolase-like
Superfamily:Purine and uridine phosphorylases
Family:Purine and uridine phosphorylases |
| 75 | d1gtma2 |
|
not modelled |
10.4 |
17 |
Fold:Aminoacid dehydrogenase-like, N-terminal domain
Superfamily:Aminoacid dehydrogenase-like, N-terminal domain
Family:Aminoacid dehydrogenases |
| 76 | c3cvoA_ |
|
not modelled |
10.3 |
15 |
PDB header:transferase
Chain: A: PDB Molecule:methyltransferase-like protein of unknown function;
PDBTitle: crystal structure of a methyltransferase-like protein (spo2022) from2 silicibacter pomeroyi dss-3 at 1.80 a resolution
|
| 77 | d1h2ka_ |
|
not modelled |
10.3 |
31 |
Fold:Double-stranded beta-helix
Superfamily:Clavaminate synthase-like
Family:Hypoxia-inducible factor HIF ihhibitor (FIH1) |
| 78 | c2cunA_ |
|
not modelled |
10.1 |
21 |
PDB header:transferase
Chain: A: PDB Molecule:phosphoglycerate kinase;
PDBTitle: crystal structure of phosphoglycerate kinase from pyrococcus2 horikoshii ot3
|
| 79 | c3k2oB_ |
|
not modelled |
9.9 |
15 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:bifunctional arginine demethylase and lysyl-hydroxylase
PDBTitle: structure of an oxygenase
|
| 80 | c2zpmA_ |
|
not modelled |
9.8 |
7 |
PDB header:hydrolase
Chain: A: PDB Molecule:regulator of sigma e protease;
PDBTitle: crystal structure analysis of pdz domain b
|
| 81 | d1n8ja_ |
|
not modelled |
9.4 |
15 |
Fold:Thioredoxin fold
Superfamily:Thioredoxin-like
Family:Glutathione peroxidase-like |
| 82 | c2x4iA_ |
|
not modelled |
9.2 |
12 |
PDB header:unknown function
Chain: A: PDB Molecule:uncharacterized protein 114;
PDBTitle: orf 114a from sulfolobus islandicus rudivirus 1
|
| 83 | c2bnoA_ |
|
not modelled |
9.1 |
10 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:epoxidase;
PDBTitle: the structure of hydroxypropylphosphonic acid epoxidase2 from s. wedmorenis.
|
| 84 | d1euza2 |
|
not modelled |
9.0 |
15 |
Fold:Aminoacid dehydrogenase-like, N-terminal domain
Superfamily:Aminoacid dehydrogenase-like, N-terminal domain
Family:Aminoacid dehydrogenases |
| 85 | d7odca1 |
|
not modelled |
8.9 |
33 |
Fold:Domain of alpha and beta subunits of F1 ATP synthase-like
Superfamily:Alanine racemase C-terminal domain-like
Family:Eukaryotic ODC-like |
| 86 | d1gph12 |
|
not modelled |
8.9 |
29 |
Fold:Ntn hydrolase-like
Superfamily:N-terminal nucleophile aminohydrolases (Ntn hydrolases)
Family:Class II glutamine amidotransferases |
| 87 | d1v4na_ |
|
not modelled |
8.3 |
10 |
Fold:Phosphorylase/hydrolase-like
Superfamily:Purine and uridine phosphorylases
Family:Purine and uridine phosphorylases |
| 88 | c2bf9A_ |
|
not modelled |
8.2 |
10 |
PDB header:hormone
Chain: A: PDB Molecule:pancreatic hormone;
PDBTitle: anisotropic refinement of avian (turkey) pancreatic2 polypeptide at 0.99 angstroms resolution.
|
| 89 | c2p1gA_ |
|
not modelled |
8.0 |
10 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:putative xylanase;
PDBTitle: crystal structure of a putative xylanase from bacteroides fragilis
|
| 90 | d1bgva2 |
|
not modelled |
8.0 |
9 |
Fold:Aminoacid dehydrogenase-like, N-terminal domain
Superfamily:Aminoacid dehydrogenase-like, N-terminal domain
Family:Aminoacid dehydrogenases |
| 91 | c3e5bB_ |
|
not modelled |
7.8 |
25 |
PDB header:lyase
Chain: B: PDB Molecule:isocitrate lyase;
PDBTitle: 2.4 a crystal structure of isocitrate lyase from brucella2 melitensis
|
| 92 | c3bvcA_ |
|
not modelled |
7.7 |
24 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:uncharacterized protein ism_01780;
PDBTitle: crystal structure of uncharacterized protein ism_01780 from2 roseovarius nubinhibens ism
|
| 93 | d1f3ta1 |
|
not modelled |
7.6 |
33 |
Fold:Domain of alpha and beta subunits of F1 ATP synthase-like
Superfamily:Alanine racemase C-terminal domain-like
Family:Eukaryotic ODC-like |
| 94 | c3khsB_ |
|
not modelled |
7.6 |
16 |
PDB header:hydrolase
Chain: B: PDB Molecule:purine nucleoside phosphorylase;
PDBTitle: crystal structure of grouper iridovirus purine nucleoside2 phosphorylase
|
| 95 | c1xjvA_ |
|
not modelled |
7.4 |
43 |
PDB header:transcription/dna
Chain: A: PDB Molecule:protection of telomeres 1;
PDBTitle: crystal structure of human pot1 bound to telomeric single-2 stranded dna (ttagggttag)
|
| 96 | c3kw0D_ |
|
not modelled |
7.4 |
20 |
PDB header:hydrolase
Chain: D: PDB Molecule:cysteine peptidase;
PDBTitle: crystal structure of cysteine peptidase (np_982244.1) from bacillus2 cereus atcc 10987 at 2.50 a resolution
|
| 97 | c3dkqB_ |
|
not modelled |
7.2 |
19 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:pkhd-type hydroxylase sbal_3634;
PDBTitle: crystal structure of putative oxygenase (yp_001051978.1) from2 shewanella baltica os155 at 2.26 a resolution
|
| 98 | c1ronA_ |
|
not modelled |
7.2 |
29 |
PDB header:neuropeptide
Chain: A: PDB Molecule:neuropeptide y;
PDBTitle: nmr solution structure of human neuropeptide y
|
| 99 | c1z7hA_ |
|
not modelled |
7.0 |
18 |
PDB header:hydrolase
Chain: A: PDB Molecule:tetanus toxin light chain;
PDBTitle: 2.3 angstrom crystal structure of tetanus neurotoxin light2 chain
|