1 c1sfeA_
100.0
32
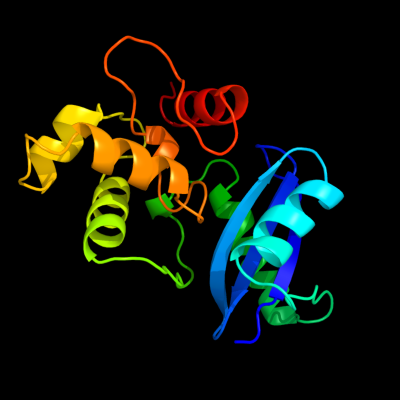
PDB header: dna-binding proteinChain: A: PDB Molecule: ada o6-methylguanine-dna methyltransferase;PDBTitle: ada o6-methylguanine-dna methyltransferase from escherichia coli
2 c1wrjA_
100.0
31
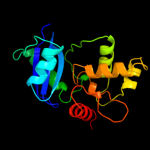
PDB header: transferaseChain: A: PDB Molecule: methylated-dna--protein-cysteinePDBTitle: crystal structure of o6-methylguanine methyltransferase2 from sulfolobus tokodaii
3 c1t39A_
100.0
35
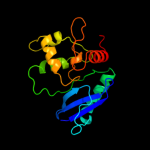
PDB header: transferase/dnaChain: A: PDB Molecule: methylated-dna--protein-cysteinePDBTitle: human o6-alkylguanine-dna alkyltransferase covalently2 crosslinked to dna
4 c1mgtA_
100.0
30
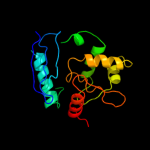
PDB header: transferaseChain: A: PDB Molecule: protein (o6-methylguanine-dna methyltransferase);PDBTitle: crystal structure of o6-methylguanine-dna methyltransferase from2 hyperthermophilic archaeon pyrococcus kodakaraensis strain kod1
5 c2g7hA_
100.0
21
PDB header: transferaseChain: A: PDB Molecule: methylated-dna--protein-cysteinePDBTitle: structure of an o6-methylguanine dna methyltransferase from2 methanococcus jannaschii (mj1529)
6 d1sfea1
100.0
52
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Methylated DNA-protein cysteine methyltransferase, C-terminal domainFamily: Methylated DNA-protein cysteine methyltransferase, C-terminal domain7 d1qnta1
100.0
50
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Methylated DNA-protein cysteine methyltransferase, C-terminal domainFamily: Methylated DNA-protein cysteine methyltransferase, C-terminal domain8 c3gx4X_
100.0
34
PDB header: dna binding protein/dnaChain: X: PDB Molecule: alkyltransferase-like protein 1;PDBTitle: crystal structure analysis of s. pombe atl in complex with dna
9 d1mgta1
100.0
44
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Methylated DNA-protein cysteine methyltransferase, C-terminal domainFamily: Methylated DNA-protein cysteine methyltransferase, C-terminal domain10 c2kimA_
100.0
31
PDB header: transferaseChain: A: PDB Molecule: o6-methylguanine-dna methyltransferase;PDBTitle: 1.7-mm microcryoprobe solution nmr structure of an o6-2 methylguanine dna methyltransferase family protein from3 vibrio parahaemolyticus. northeast structural genomics4 consortium target vpr247.
11 d1sfea2
99.0
12
Fold: Ribonuclease H-like motifSuperfamily: Methylated DNA-protein cysteine methyltransferase domainFamily: Methylated DNA-protein cysteine methyltransferase domain12 d1qnta2
96.2
18
Fold: Ribonuclease H-like motifSuperfamily: Methylated DNA-protein cysteine methyltransferase domainFamily: Methylated DNA-protein cysteine methyltransferase domain13 c1dpuA_
68.8
19
PDB header: dna binding proteinChain: A: PDB Molecule: replication protein a (rpa32) c-terminal domain;PDBTitle: solution structure of the c-terminal domain of human rpa322 complexed with ung2(73-88)
14 d1dpua_
68.8
19
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: C-terminal domain of RPA3215 d2gxba1
64.6
23
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: Z-DNA binding domain16 d1qgpa_
58.3
22
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: Z-DNA binding domain17 d1stza1
57.8
11
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: Heat-inducible transcription repressor HrcA, N-terminal domain18 d1qbjc_
54.2
23
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: Z-DNA binding domain19 d1jhfa1
40.7
22
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: LexA repressor, N-terminal DNA-binding domain20 c3k69A_
37.9
19
PDB header: transcriptionChain: A: PDB Molecule: putative transcription regulator;PDBTitle: crystal structure of a putative transcriptional regulator (lp_0360)2 from lactobacillus plantarum at 1.95 a resolution
21 d1tnsa_
not modelled
35.3
22
Fold: Putative DNA-binding domainSuperfamily: Putative DNA-binding domainFamily: Excisionase-like22 c3lwfD_
not modelled
33.1
16
PDB header: transcription regulatorChain: D: PDB Molecule: putative transcriptional regulator;PDBTitle: crystal structure of putative transcriptional regulator (np_470886.1)2 from listeria innocua at 2.06 a resolution
23 d1cf7b_
not modelled
23.1
21
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: Cell cycle transcription factor e2f-dp24 c2l01A_
not modelled
22.8
10
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: uncharacterized protein;PDBTitle: solution nmr structure of protein bvu3908 from bacteroides vulgatus,2 northeast structural genomics consortium target bvr153
25 d2cyya1
not modelled
21.7
10
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: Lrp/AsnC-like transcriptional regulator N-terminal domain26 d1bw6a_
not modelled
20.5
11
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Homeodomain-likeFamily: Centromere-binding27 c3c8lB_
not modelled
19.9
40
PDB header: unknown functionChain: B: PDB Molecule: ftsz-like protein of unknown function;PDBTitle: crystal structure of a ftsz-like protein of unknown function2 (npun_r1471) from nostoc punctiforme pcc 73102 at 1.22 a resolution
28 d1ub9a_
not modelled
17.8
15
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: MarR-like transcriptional regulators29 d1or7a1
not modelled
17.6
15
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Sigma3 and sigma4 domains of RNA polymerase sigma factorsFamily: Sigma4 domain30 c2l02B_
not modelled
17.4
27
PDB header: structural genomics, unknown functionChain: B: PDB Molecule: uncharacterized protein;PDBTitle: solution nmr structure of protein bt2368 from bacteroides2 thetaiotaomicron, northeast structural genomics consortium target3 btr375
31 c2o8xA_
not modelled
17.2
14
PDB header: transcriptionChain: A: PDB Molecule: probable rna polymerase sigma-c factor;PDBTitle: crystal structure of the "-35 element" promoter recognition domain of2 mycobacterium tuberculosis sigc
32 c3k2zA_
not modelled
16.8
18
PDB header: hydrolaseChain: A: PDB Molecule: lexa repressor;PDBTitle: crystal structure of a lexa protein from thermotoga maritima
33 d1u0ma2
not modelled
16.7
13
Fold: Thiolase-likeSuperfamily: Thiolase-likeFamily: Chalcone synthase-like34 d1mkma1
not modelled
15.8
14
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: Transcriptional regulator IclR, N-terminal domain35 c2cfxD_
not modelled
15.6
7
PDB header: transcriptionChain: D: PDB Molecule: hth-type transcriptional regulator lrpc;PDBTitle: structure of b.subtilis lrpc
36 c3g5oA_
not modelled
15.2
13
PDB header: toxin/antitoxinChain: A: PDB Molecule: uncharacterized protein rv2865;PDBTitle: the crystal structure of the toxin-antitoxin complex relbe2 (rv2865-2 2866) from mycobacterium tuberculosis
37 c3fvcA_
not modelled
14.2
29
PDB header: viral proteinChain: A: PDB Molecule: glycoprotein gp110;PDBTitle: crystal structure of a trimeric variant of the epstein-barr virus2 glycoprotein b
38 c2iv1J_
not modelled
14.1
16
PDB header: lyaseChain: J: PDB Molecule: cyanate hydratase;PDBTitle: site directed mutagenesis of key residues involved in the2 catalytic mechanism of cyanase
39 d1dwka1
not modelled
12.7
16
Fold: lambda repressor-like DNA-binding domainsSuperfamily: lambda repressor-like DNA-binding domainsFamily: Cyanase N-terminal domain40 d2cfxa1
not modelled
12.3
7
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: Lrp/AsnC-like transcriptional regulator N-terminal domain41 d1ku7a_
not modelled
12.3
10
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Sigma3 and sigma4 domains of RNA polymerase sigma factorsFamily: Sigma4 domain42 c2da5A_
not modelled
12.3
13
PDB header: dna binding proteinChain: A: PDB Molecule: zinc fingers and homeoboxes protein 3;PDBTitle: solution structure of the second homeobox domain of zinc2 fingers and homeoboxes protein 3 (triple homeobox 13 protein)
43 c3r0aB_
not modelled
12.2
18
PDB header: transcription regulatorChain: B: PDB Molecule: putative transcriptional regulator;PDBTitle: possible transcriptional regulator from methanosarcina mazei go1 (gi2 21227196)
44 c3hugA_
not modelled
12.2
17
PDB header: transcription/membrane proteinChain: A: PDB Molecule: rna polymerase sigma factor;PDBTitle: crystal structure of mycobacterium tuberculosis anti-sigma factor rsla2 in complex with -35 promoter binding domain of sigl
45 d1s6la1
not modelled
11.9
21
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: MerB N-terminal domain-like46 c3t72o_
not modelled
11.6
13
PDB header: transcription/dnaChain: O: PDB Molecule: pho box dna (strand 1);PDBTitle: phob(e)-sigma70(4)-(rnap-betha-flap-tip-helix)-dna transcription2 activation sub-complex
47 c2dbbA_
not modelled
11.5
15
PDB header: transcriptional regulatorChain: A: PDB Molecule: putative hth-type transcriptional regulator ph0061;PDBTitle: crystal structure of ph0061
48 d1s7oa_
not modelled
11.5
18
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Sigma3 and sigma4 domains of RNA polymerase sigma factorsFamily: YlxM/p13-like49 d1ku3a_
not modelled
11.3
11
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Sigma3 and sigma4 domains of RNA polymerase sigma factorsFamily: Sigma4 domain50 c2e1cA_
not modelled
11.1
10
PDB header: transcription/dnaChain: A: PDB Molecule: putative hth-type transcriptional regulator ph1519;PDBTitle: structure of putative hth-type transcriptional regulator ph1519/dna2 complex
51 d1dhsa_
not modelled
11.0
18
Fold: DHS-like NAD/FAD-binding domainSuperfamily: DHS-like NAD/FAD-binding domainFamily: Deoxyhypusine synthase, DHS52 d1biaa1
not modelled
10.9
19
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: Biotin repressor-like53 c1mkmA_
not modelled
10.2
14
PDB header: transcriptionChain: A: PDB Molecule: iclr transcriptional regulator;PDBTitle: crystal structure of the thermotoga maritima iclr
54 c1i1gA_
not modelled
9.7
15
PDB header: transcriptionChain: A: PDB Molecule: transcriptional regulator lrpa;PDBTitle: crystal structure of the lrp-like transcriptional regulator from the2 archaeon pyrococcus furiosus
55 c3narA_
not modelled
9.4
25
PDB header: transcriptionChain: A: PDB Molecule: zinc fingers and homeoboxes protein 1;PDBTitle: crystal structure of zhx1 hd4 (zinc-fingers and homeoboxes protein 1,2 homeodomain 4)
56 c2y75F_
not modelled
9.2
10
PDB header: transcriptionChain: F: PDB Molecule: hth-type transcriptional regulator cymr;PDBTitle: the structure of cymr (yrzc) the global cysteine regulator2 of b. subtilis
57 d1rp3a2
not modelled
9.2
11
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Sigma3 and sigma4 domains of RNA polymerase sigma factorsFamily: Sigma4 domain58 d1xmka1
not modelled
9.1
19
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: Z-DNA binding domain59 d2guma1
not modelled
8.9
20
Fold: Viral glycoprotein ectodomain-likeSuperfamily: Viral glycoprotein ectodomain-likeFamily: Glycoprotein B-like60 c3nw8B_
not modelled
8.8
20
PDB header: viral proteinChain: B: PDB Molecule: envelope glycoprotein b;PDBTitle: glycoprotein b from herpes simplex virus type 1, y179s mutant, high-ph
61 c2inpD_
not modelled
8.7
38
PDB header: oxidoreductaseChain: D: PDB Molecule: phenol hydroxylase component phl;PDBTitle: structure of the phenol hydroxylase-regulatory protein2 complex
62 c3i4pA_
not modelled
8.7
13
PDB header: transcription regulatorChain: A: PDB Molecule: transcriptional regulator, asnc family;PDBTitle: crystal structure of asnc family transcriptional regulator from2 agrobacterium tumefaciens
63 d2cg4a1
not modelled
8.6
18
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: Lrp/AsnC-like transcriptional regulator N-terminal domain64 d2f3ci1
not modelled
8.6
56
Fold: Kazal-type serine protease inhibitorsSuperfamily: Kazal-type serine protease inhibitorsFamily: Ovomucoid domain III-like65 d2p7vb1
not modelled
8.5
10
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Sigma3 and sigma4 domains of RNA polymerase sigma factorsFamily: Sigma4 domain66 d2hs5a1
not modelled
8.4
21
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: GntR-like transcriptional regulators67 d1xsva_
not modelled
8.4
14
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Sigma3 and sigma4 domains of RNA polymerase sigma factorsFamily: YlxM/p13-like68 c2ia0A_
not modelled
8.3
15
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: putative hth-type transcriptional regulator pf0864;PDBTitle: transcriptional regulatory protein pf0864 from pyrococcus furiosus a2 member of the asnc family (pf0864)
69 d1wh5a_
not modelled
8.3
21
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Homeodomain-likeFamily: Homeodomain70 d1s7ea1
not modelled
8.0
19
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Homeodomain-likeFamily: Homeodomain71 c3ee6A_
not modelled
7.9
10
PDB header: hydrolaseChain: A: PDB Molecule: tripeptidyl-peptidase 1;PDBTitle: crystal structure analysis of tripeptidyl peptidase -i
72 d1i1ga1
not modelled
7.8
15
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: Lrp/AsnC-like transcriptional regulator N-terminal domain73 d3ctaa1
not modelled
7.5
17
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: MarR-like transcriptional regulators74 d2ecba1
not modelled
7.5
16
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Homeodomain-likeFamily: Homeodomain75 d2hi3a1
not modelled
7.5
16
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Homeodomain-likeFamily: Homeodomain76 d1ku2a1
not modelled
7.4
24
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Sigma3 and sigma4 domains of RNA polymerase sigma factorsFamily: Sigma3 domain77 d1xd7a_
not modelled
7.4
14
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: Transcriptional regulator Rrf278 c1rr7A_
not modelled
7.4
11
PDB header: transcriptionChain: A: PDB Molecule: middle operon regulator;PDBTitle: crystal structure of the middle operon regulator protein of2 bacteriophage mu
79 d1rr7a_
not modelled
7.4
11
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Homeodomain-likeFamily: Middle operon regulator, Mor80 d1smyf2
not modelled
7.3
8
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Sigma3 and sigma4 domains of RNA polymerase sigma factorsFamily: Sigma4 domain81 c3qmlC_
not modelled
7.3
21
PDB header: chaperone/protein transportChain: C: PDB Molecule: nucleotide exchange factor sil1;PDBTitle: the structural analysis of sil1-bip complex reveals the mechanism for2 sil1 to function as a novel nucleotide exchange factor
82 d1nffa_
not modelled
7.2
11
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: Tyrosine-dependent oxidoreductases83 c1r22B_
not modelled
7.2
22
PDB header: transcription repressorChain: B: PDB Molecule: transcriptional repressor smtb;PDBTitle: crystal structure of the cyanobacterial metallothionein2 repressor smtb (c14s/c61s/c121s mutant) in the zn2alpha5-3 form
84 d1ttya_
not modelled
7.2
7
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Sigma3 and sigma4 domains of RNA polymerase sigma factorsFamily: Sigma4 domain85 c2e7xA_
not modelled
7.2
10
PDB header: transcription regulatorChain: A: PDB Molecule: 150aa long hypothetical transcriptional regulator;PDBTitle: structure of the lrp/asnc like transcriptional regulator from2 sulfolobus tokodaii 7 complexed with its cognate ligand
86 c2l4aA_
not modelled
7.1
10
PDB header: dna binding proteinChain: A: PDB Molecule: leucine responsive regulatory protein;PDBTitle: nmr structure of the dna-binding domain of e.coli lrp
87 d1ig6a_
not modelled
6.9
8
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: ARID-likeFamily: ARID domain88 c3eusB_
not modelled
6.8
26
PDB header: dna binding proteinChain: B: PDB Molecule: dna-binding protein;PDBTitle: the crystal structure of the dna binding protein from silicibacter2 pomeroyi
89 c3a01A_
not modelled
6.8
16
PDB header: gene regulation/dnaChain: A: PDB Molecule: homeodomain-containing protein;PDBTitle: crystal structure of aristaless and clawless homeodomains bound to dna
90 c3sztB_
not modelled
6.7
17
PDB header: transcriptionChain: B: PDB Molecule: quorum-sensing control repressor;PDBTitle: quorum sensing control repressor, qscr, bound to n-3-oxo-dodecanoyl-l-2 homoserine lactone
91 c2kxxA_
not modelled
6.6
36
PDB header: protein bindingChain: A: PDB Molecule: small protein a;PDBTitle: nmr structure of escherichia coli bame, a lipoprotein component of the2 beta-barrel assembly machinery complex
92 d1t1ea2
not modelled
6.4
6
Fold: Ferredoxin-likeSuperfamily: Protease propeptides/inhibitorsFamily: Subtilase propeptides/inhibitors93 d1hlva1
not modelled
6.4
11
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Homeodomain-likeFamily: Centromere-binding94 d1rg6a_
not modelled
6.4
21
Fold: SAM domain-likeSuperfamily: SAM/Pointed domainFamily: SAM (sterile alpha motif) domain95 d1p2fa1
not modelled
6.3
20
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: C-terminal effector domain of the bipartite response regulatorsFamily: PhoB-like96 c1x3uA_
not modelled
6.1
16
PDB header: transcriptionChain: A: PDB Molecule: transcriptional regulatory protein fixj;PDBTitle: solution structure of the c-terminal transcriptional2 activator domain of fixj from sinorhizobium melilot
97 c3frwF_
not modelled
6.0
14
PDB header: structural genomics, unknown functionChain: F: PDB Molecule: putative trp repressor protein;PDBTitle: crystal structure of putative trpr protein from ruminococcus obeum
98 c2oqgA_
not modelled
6.0
19
PDB header: transcriptionChain: A: PDB Molecule: possible transcriptional regulator, arsr family protein;PDBTitle: arsr-like transcriptional regulator from rhodococcus sp. rha1
99 c3mq0A_
not modelled
6.0
13
PDB header: transcription repressorChain: A: PDB Molecule: transcriptional repressor of the blcabc operon;PDBTitle: crystal structure of agobacterium tumefaciens repressor blcr