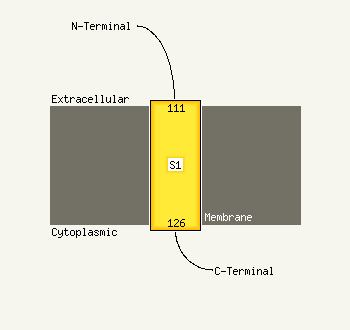
1 d1jpya_
51.2
15
Fold: Cystine-knot cytokinesSuperfamily: Cystine-knot cytokinesFamily: Interleukin 17F, IL-17F2 c2z0bB_
39.7
21
PDB header: hydrolaseChain: B: PDB Molecule: putative glycerophosphodiester phosphodiesterase 5;PDBTitle: crystal structure of cbm20 domain of human putative2 glycerophosphodiester phosphodiesterase 5 (kiaa1434)
3 c2kv5A_
25.5
29
PDB header: toxinChain: A: PDB Molecule: putative uncharacterized protein rnai;PDBTitle: solution structure of the par toxin fst in dpc micelles
4 c2wskA_
14.8
25
PDB header: hydrolaseChain: A: PDB Molecule: glycogen debranching enzyme;PDBTitle: crystal structure of glycogen debranching enzyme glgx from2 escherichia coli k-12
5 d2zkmx4
13.7
55
Fold: TIM beta/alpha-barrelSuperfamily: PLC-like phosphodiesterasesFamily: Mammalian PLC6 d1qasa3
13.5
45
Fold: TIM beta/alpha-barrelSuperfamily: PLC-like phosphodiesterasesFamily: Mammalian PLC7 d1cyga2
13.2
14
Fold: Prealbumin-likeSuperfamily: Starch-binding domain-likeFamily: Starch-binding domain8 d1kpta_
13.0
38
Fold: Yeast killer toxinsSuperfamily: Yeast killer toxinsFamily: Virally encoded KP4 toxin9 c3cdzA_
11.2
10
PDB header: blood clottingChain: A: PDB Molecule: coagulation factor viii heavy chain;PDBTitle: crystal structure of human factor viii
10 d3bmva2
10.8
19
Fold: Prealbumin-likeSuperfamily: Starch-binding domain-likeFamily: Starch-binding domain11 d1a73a_
10.4
21
Fold: His-Me finger endonucleasesSuperfamily: His-Me finger endonucleasesFamily: Intron-encoded homing endonucleases12 c3spaA_
10.2
17
PDB header: transferaseChain: A: PDB Molecule: dna-directed rna polymerase, mitochondrial;PDBTitle: crystal structure of human mitochondrial rna polymerase
13 c3pnrB_
8.8
56
PDB header: hydrolase/hydrolase inhibitorChain: B: PDB Molecule: pbicp-c;PDBTitle: structure of pbicp-c in complex with falcipain-2
14 d1iz5a2
8.3
21
Fold: DNA clampSuperfamily: DNA clampFamily: DNA polymerase processivity factor15 d1kula_
7.8
15
Fold: Prealbumin-likeSuperfamily: Starch-binding domain-likeFamily: Starch-binding domain16 c3l81A_
7.7
13
PDB header: transport proteinChain: A: PDB Molecule: ap-4 complex subunit mu-1;PDBTitle: crystal structure of adaptor protein complex 4 (ap-4) mu4 subunit c-2 terminal domain, in complex with a sorting peptide from the amyloid3 precursor protein (app)
17 c1djyB_
7.6
45
PDB header: lipid degradationChain: B: PDB Molecule: phosphoinositide-specific phospholipase c,PDBTitle: phosphoinositide-specific phospholipase c-delta1 from rat2 complexed with inositol-2,4,5-trisphosphate
18 d1ud9a2
7.5
26
Fold: DNA clampSuperfamily: DNA clampFamily: DNA polymerase processivity factor19 c2ix2A_
7.1
20
PDB header: replicationChain: A: PDB Molecule: dna polymerase sliding clamp b;PDBTitle: crystal structure of the heterotrimeric pcna from2 sulfolobus solfataricus
20 d1mswd_
7.0
21
Fold: DNA/RNA polymerasesSuperfamily: DNA/RNA polymerasesFamily: T7 RNA polymerase21 c3tu8A_
not modelled
7.0
31
PDB header: unknown functionChain: A: PDB Molecule: burkholderia lethal factor 1 (blf1);PDBTitle: crystal structure of the burkholderia lethal factor 1 (blf1)
22 d1u7ba2
not modelled
6.8
26
Fold: DNA clampSuperfamily: DNA clampFamily: DNA polymerase processivity factor23 c2k1iA_
not modelled
6.2
47
PDB header: antimicrobial proteinChain: A: PDB Molecule: mucosal alpha-defensin;PDBTitle: synthesis, structure and activities of an oral mucosal2 alpha-defensin from rhesus macaque
24 d1fx0b2
not modelled
6.2
26
Fold: Domain of alpha and beta subunits of F1 ATP synthase-likeSuperfamily: N-terminal domain of alpha and beta subunits of F1 ATP synthaseFamily: N-terminal domain of alpha and beta subunits of F1 ATP synthase25 d2jdid2
not modelled
6.0
17
Fold: Domain of alpha and beta subunits of F1 ATP synthase-likeSuperfamily: N-terminal domain of alpha and beta subunits of F1 ATP synthaseFamily: N-terminal domain of alpha and beta subunits of F1 ATP synthase26 c3ohmB_
not modelled
6.0
60
PDB header: signaling protein / hydrolaseChain: B: PDB Molecule: 1-phosphatidylinositol-4,5-bisphosphate phosphodiesterasePDBTitle: crystal structure of activated g alpha q bound to its effector2 phospholipase c beta 3
27 c2fjuB_
not modelled
5.9
60
PDB header: signaling protein,apoptosis/hydrolaseChain: B: PDB Molecule: 1-phosphatidylinositol-4,5-bisphosphatePDBTitle: activated rac1 bound to its effector phospholipase c beta 2
28 d1cxla2
not modelled
5.6
14
Fold: Prealbumin-likeSuperfamily: Starch-binding domain-likeFamily: Starch-binding domain29 d1gpla1
not modelled
5.5
26
Fold: Lipase/lipooxygenase domain (PLAT/LH2 domain)Superfamily: Lipase/lipooxygenase domain (PLAT/LH2 domain)Family: Colipase-binding domain30 d1wb1a1
not modelled
5.4
11
Fold: Reductase/isomerase/elongation factor common domainSuperfamily: Translation proteinsFamily: Elongation factors31 d1mabb2
not modelled
5.3
17
Fold: Domain of alpha and beta subunits of F1 ATP synthase-likeSuperfamily: N-terminal domain of alpha and beta subunits of F1 ATP synthaseFamily: N-terminal domain of alpha and beta subunits of F1 ATP synthase32 d1edqa1
not modelled
5.3
16
Fold: Immunoglobulin-like beta-sandwichSuperfamily: E set domainsFamily: E-set domains of sugar-utilizing enzymes