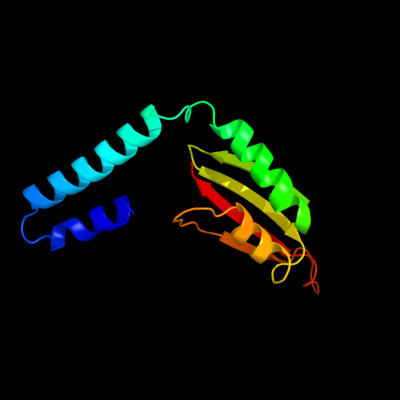
1 c2rjzA_
96.9
10
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: pilo protein;PDBTitle: crystal structure of the type 4 fimbrial biogenesis protein pilo from2 pseudomonas aeruginosa
2 c2w7vB_
61.4
17
PDB header: transport proteinChain: B: PDB Molecule: general secretion pathway protein l;PDBTitle: periplasmic domain of epsl from vibrio parahaemolyticus
3 c3mc8A_
49.0
6
PDB header: membrane proteinChain: A: PDB Molecule: alr2269 protein;PDBTitle: potra1-3 of the periplasmic domain of omp85 from anabaena
4 c2x8xX_
27.2
13
PDB header: chaperoneChain: X: PDB Molecule: tlr1789 protein;PDBTitle: structure of the n-terminal domain of omp85 from the2 thermophilic cyanobacterium thermosynechococcus elongatus
5 c2kgsA_
26.3
29
PDB header: membrane proteinChain: A: PDB Molecule: uncharacterized protein rv0899/mt0922;PDBTitle: solution structure of the amino-terminal domain of ompatb, a pore2 forming protein from mycobacterium tuberculosis
6 c2j8pA_
23.3
26
PDB header: nuclear proteinChain: A: PDB Molecule: cleavage stimulation factor 64 kda subunit;PDBTitle: nmr structure of c-terminal domain of human cstf-64
7 d1w2ia_
23.0
14
Fold: Ferredoxin-likeSuperfamily: Acylphosphatase/BLUF domain-likeFamily: Acylphosphatase-like8 c1uv7A_
22.8
10
PDB header: transportChain: A: PDB Molecule: general secretion pathway protein m;PDBTitle: periplasmic domain of epsm from vibrio cholerae
9 d1uv7a_
22.8
10
Fold: RRF/tRNA synthetase additional domain-likeSuperfamily: General secretion pathway protein M, EpsMFamily: General secretion pathway protein M, EpsM10 c2kxyA_
19.2
19
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: uncharacterized protein;PDBTitle: solution structure of sur18c from streptococcus thermophilus.2 northeast structural genomics consortium target sur18c
11 c3og5A_
17.3
9
PDB header: protein bindingChain: A: PDB Molecule: outer membrane protein assembly complex, yaet protein;PDBTitle: crystal structure of bama potra45 tandem
12 c3hd7A_
16.0
12
PDB header: exocytosisChain: A: PDB Molecule: vesicle-associated membrane protein 2;PDBTitle: helical extension of the neuronal snare complex into the membrane,2 spacegroup c 1 2 1
13 c3br8A_
14.7
8
PDB header: hydrolaseChain: A: PDB Molecule: probable acylphosphatase;PDBTitle: crystal structure of acylphosphatase from bacillus subtilis
14 c3ibwA_
14.1
13
PDB header: transferaseChain: A: PDB Molecule: gtp pyrophosphokinase;PDBTitle: crystal structure of the act domain from gtp2 pyrophosphokinase of chlorobium tepidum. northeast3 structural genomics consortium target ctr148a
15 c3qq5A_
13.4
33
PDB header: oxidoreductaseChain: A: PDB Molecule: small gtp-binding protein;PDBTitle: crystal structure of the [fefe]-hydrogenase maturation protein hydf
16 c3k2kA_
13.1
17
PDB header: hydrolaseChain: A: PDB Molecule: putative carboxypeptidase;PDBTitle: crystal structure of putative carboxypeptidase (yp_103406.1) from2 burkholderia mallei atcc 23344 at 2.49 a resolution
17 d1ulra_
13.0
15
Fold: Ferredoxin-likeSuperfamily: Acylphosphatase/BLUF domain-likeFamily: Acylphosphatase-like18 d1gnwa1
11.8
9
Fold: GST C-terminal domain-likeSuperfamily: GST C-terminal domain-likeFamily: Glutathione S-transferase (GST), C-terminal domain19 d2enda_
11.0
7
Fold: T4 endonuclease VSuperfamily: T4 endonuclease VFamily: T4 endonuclease V20 c2hsmA_
10.9
18
PDB header: ligase/rna binding proteinChain: A: PDB Molecule: glutamyl-trna synthetase, cytoplasmic;PDBTitle: structural basis of yeast aminoacyl-trna synthetase complex2 formation revealed by crystal structures of two binary sub-3 complexes
21 d1pmta1
not modelled
10.8
13
Fold: GST C-terminal domain-likeSuperfamily: GST C-terminal domain-likeFamily: Glutathione S-transferase (GST), C-terminal domain22 d2awna1
not modelled
10.4
63
Fold: OB-foldSuperfamily: MOP-likeFamily: ABC-transporter additional domain23 c1qcrD_
not modelled
10.1
11
PDB header: PDB COMPND: 24 c2x3dC_
not modelled
9.6
11
PDB header: unknown functionChain: C: PDB Molecule: sso6206;PDBTitle: crystal structure of sso6206 from sulfolobus solfataricus p2
25 c3lygA_
not modelled
9.5
28
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: ntf2-like protein of unknown function;PDBTitle: crystal structure of ntf2-like protein of unknown function2 (yp_270605.1) from colwellia psychrerythraea 34h at 1.61 a resolution
26 d3bzfa1
not modelled
8.9
10
Fold: Immunoglobulin-like beta-sandwichSuperfamily: ImmunoglobulinFamily: C1 set domains (antibody constant domain-like)27 d1vm0a_
not modelled
8.8
12
Fold: IF3-likeSuperfamily: AlbA-likeFamily: Hypothetical protein At2g3416028 d1dx4a_
not modelled
8.2
11
Fold: alpha/beta-HydrolasesSuperfamily: alpha/beta-HydrolasesFamily: Acetylcholinesterase-like29 c1f8uA_
not modelled
8.0
11
PDB header: hydrolase/toxinChain: A: PDB Molecule: acetylcholinesterase;PDBTitle: crystal structure of mutant e202q of human acetylcholinesterase2 complexed with green mamba venom peptide fasciculin-ii
30 d1f8ua_
not modelled
8.0
11
Fold: alpha/beta-HydrolasesSuperfamily: alpha/beta-HydrolasesFamily: Acetylcholinesterase-like31 c1p84D_
not modelled
7.9
16
PDB header: oxidoreductaseChain: D: PDB Molecule: cytochrome c1, heme protein;PDBTitle: hdbt inhibited yeast cytochrome bc1 complex
32 d2ha2a1
not modelled
7.6
14
Fold: alpha/beta-HydrolasesSuperfamily: alpha/beta-HydrolasesFamily: Acetylcholinesterase-like33 d1rl2a1
not modelled
7.5
18
Fold: SH3-like barrelSuperfamily: Translation proteins SH3-like domainFamily: C-terminal domain of ribosomal protein L234 c3ipfA_
not modelled
7.4
25
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: uncharacterized protein;PDBTitle: crystal structure of the q251q8_deshy protein from desulfitobacterium2 hafniense. northeast structural genomics consortium target dhr8c.
35 d1qo3a1
not modelled
7.4
13
Fold: Immunoglobulin-like beta-sandwichSuperfamily: ImmunoglobulinFamily: C1 set domains (antibody constant domain-like)36 c2l5gB_
not modelled
7.4
13
PDB header: transcription regulatorChain: B: PDB Molecule: putative uncharacterized protein ncor2;PDBTitle: co-ordinates and 1h, 13c and 15n chemical shift assignments for the2 complex of gps2 53-90 and smrt 167-207
37 d1ea5a_
not modelled
7.3
14
Fold: alpha/beta-HydrolasesSuperfamily: alpha/beta-HydrolasesFamily: Acetylcholinesterase-like38 d2cyya2
not modelled
7.3
14
Fold: Ferredoxin-likeSuperfamily: Dimeric alpha+beta barrelFamily: Lrp/AsnC-like transcriptional regulator C-terminal domain39 c2akrA_
not modelled
7.3
19
PDB header: immune systemChain: A: PDB Molecule: t-cell surface glycoprotein cd1d1;PDBTitle: structural basis of sulfatide presentation by mouse cd1d
40 d1onqa1
not modelled
7.2
18
Fold: Immunoglobulin-like beta-sandwichSuperfamily: ImmunoglobulinFamily: C1 set domains (antibody constant domain-like)41 c2l26A_
not modelled
7.1
29
PDB header: membrane proteinChain: A: PDB Molecule: uncharacterized protein rv0899/mt0922;PDBTitle: rv0899 from mycobacterium tuberculosis contains two separated domains
42 c2jsxA_
not modelled
6.9
17
PDB header: chaperoneChain: A: PDB Molecule: protein napd;PDBTitle: solution structure of the e. coli tat proofreading2 chaperone protein napd
43 c1xz0A_
not modelled
6.8
19
PDB header: immune systemChain: A: PDB Molecule: t-cell surface glycoprotein cd1a;PDBTitle: crystal structure of cd1a in complex with a synthetic mycobactin2 lipopeptide
44 d1f6ga_
not modelled
6.8
7
Fold: Voltage-gated potassium channelsSuperfamily: Voltage-gated potassium channelsFamily: Voltage-gated potassium channels45 c3af5A_
not modelled
6.7
17
PDB header: hydrolaseChain: A: PDB Molecule: putative uncharacterized protein ph1404;PDBTitle: the crystal structure of an archaeal cpsf subunit, ph1404 from2 pyrococcus horikoshii
46 c1cpbA_
not modelled
6.6
13
PDB header: hydrolase (c-terminal peptidase)Chain: A: PDB Molecule: carboxypeptidase b;PDBTitle: structure of carboxypeptidase b at 2.8 angstroms resolution
47 d2raqa1
not modelled
6.5
15
Fold: Ferredoxin-likeSuperfamily: MTH889-likeFamily: MTH889-like48 d1n2aa1
not modelled
6.5
5
Fold: GST C-terminal domain-likeSuperfamily: GST C-terminal domain-likeFamily: Glutathione S-transferase (GST), C-terminal domain49 c1kdxB_
not modelled
6.4
28
PDB header: transcription regulation complexChain: B: PDB Molecule: creb;PDBTitle: kix domain of mouse cbp (creb binding protein) in complex2 with phosphorylated kinase inducible domain (pkid) of rat3 creb (cyclic amp response element binding protein), nmr 174 structures
50 d1j5ya2
not modelled
6.3
8
Fold: HPr-likeSuperfamily: Putative transcriptional regulator TM1602, C-terminal domainFamily: Putative transcriptional regulator TM1602, C-terminal domain51 d2cqla1
not modelled
6.1
39
Fold: Ribosomal protein L6Superfamily: Ribosomal protein L6Family: Ribosomal protein L652 d2axtj1
not modelled
6.1
18
Fold: Single transmembrane helixSuperfamily: Photosystem II reaction center protein J, PsbJFamily: PsbJ-like53 c2uvaI_
not modelled
6.1
22
PDB header: transferaseChain: I: PDB Molecule: fatty acid synthase beta subunits;PDBTitle: crystal structure of fatty acid synthase from thermomyces2 lanuginosus at 3.1 angstrom resolution. this file contains3 the beta subunits of the fatty acid synthase. the entire4 crystal structure consists of one heterododecameric fatty5 acid synthase and is described in remark 400
54 d1k4ya_
not modelled
6.0
19
Fold: alpha/beta-HydrolasesSuperfamily: alpha/beta-HydrolasesFamily: Acetylcholinesterase-like55 c3l2nA_
not modelled
6.0
13
PDB header: hydrolaseChain: A: PDB Molecule: peptidase m14, carboxypeptidase a;PDBTitle: crystal structure of putative carboxypeptidase a (yp_562911.1) from2 shewanella denitrificans os-217 at 2.39 a resolution
56 d3bpda1
not modelled
6.0
11
Fold: Ferredoxin-likeSuperfamily: MTH889-likeFamily: MTH889-like57 c1qysA_
not modelled
5.8
15
PDB header: de novo proteinChain: A: PDB Molecule: top7;PDBTitle: crystal structure of top7: a computationally designed2 protein with a novel fold
58 d1p0ia_
not modelled
5.7
8
Fold: alpha/beta-HydrolasesSuperfamily: alpha/beta-HydrolasesFamily: Acetylcholinesterase-like59 c2e0gA_
not modelled
5.7
32
PDB header: replicationChain: A: PDB Molecule: chromosomal replication initiator protein dnaa;PDBTitle: dnaa n-terminal domain
60 c2pm8A_
not modelled
5.6
8
PDB header: hydrolaseChain: A: PDB Molecule: cholinesterase;PDBTitle: crystal structure of recombinant full length human2 butyrylcholinesterase
61 c1gzpA_
not modelled
5.6
15
PDB header: glycoproteinChain: A: PDB Molecule: t-cell surface glycoprotein cd1b;PDBTitle: cd1b in complex with gm2 ganglioside
62 c2qczA_
not modelled
5.5
10
PDB header: membrane protein, protein transportChain: A: PDB Molecule: outer membrane protein assembly factor yaet;PDBTitle: structure of n-terminal domain of e. coli yaet
63 d1rl6a1
not modelled
5.5
33
Fold: Ribosomal protein L6Superfamily: Ribosomal protein L6Family: Ribosomal protein L664 c2uz8A_
not modelled
5.5
0
PDB header: rna-binding proteinChain: A: PDB Molecule: eukaryotic translation elongation factor 1PDBTitle: the crystal structure of p18, human translation elongation2 factor 1 epsilon 1
65 c1cd1C_
not modelled
5.5
16
PDB header: cd1Chain: C: PDB Molecule: cd1;PDBTitle: cd1(mouse) antigen presenting molecule
66 c2xr1B_
not modelled
5.4
14
PDB header: hydrolaseChain: B: PDB Molecule: cleavage and polyadenylation specificity factor 100 kdPDBTitle: dimeric archaeal cleavage and polyadenylation specificity2 factor with n-terminal kh domains (kh-cpsf) from methanosarcina3 mazei
67 d1qe3a_
not modelled
5.4
11
Fold: alpha/beta-HydrolasesSuperfamily: alpha/beta-HydrolasesFamily: Acetylcholinesterase-like68 c1gnwA_
not modelled
5.4
9
PDB header: transferaseChain: A: PDB Molecule: glutathione s-transferase;PDBTitle: structure of glutathione s-transferase
69 d2vjva1
not modelled
5.4
3
Fold: Ferredoxin-likeSuperfamily: Transposase IS200-likeFamily: Transposase IS200-like70 c2vhgB_
not modelled
5.3
3
PDB header: dna-binding proteinChain: B: PDB Molecule: transposase orfa;PDBTitle: crystal structure of the ishp608 transposase in complex2 with right end 31-mer dna
71 d2zjre2
not modelled
5.3
28
Fold: Ribosomal protein L6Superfamily: Ribosomal protein L6Family: Ribosomal protein L672 c1t7wA_
not modelled
5.3
14
PDB header: lipid binding proteinChain: A: PDB Molecule: zinc-alpha-2-glycoprotein;PDBTitle: zn-alpha-2-glycoprotein; cho-zag peg 400
73 c2kncA_
not modelled
5.2
17
PDB header: cell adhesionChain: A: PDB Molecule: integrin alpha-iib;PDBTitle: platelet integrin alfaiib-beta3 transmembrane-cytoplasmic2 heterocomplex
74 c2ogsA_
not modelled
5.2
11
PDB header: hydrolaseChain: A: PDB Molecule: thermostable carboxylesterase est50;PDBTitle: crystal structure of the geobacillus stearothermophilus2 carboxylesterase est55 at ph 6.2
75 c2gv1A_
not modelled
5.2
9
PDB header: hydrolaseChain: A: PDB Molecule: probable acylphosphatase;PDBTitle: nmr solution structure of the acylphosphatase from2 eschaerichia coli
76 c3efcA_
not modelled
5.1
10
PDB header: membrane proteinChain: A: PDB Molecule: outer membrane protein assembly factor yaet;PDBTitle: crystal structure of yaet periplasmic domain
77 c2zxxA_
not modelled
5.1
13
PDB header: cell cycle/replicationChain: A: PDB Molecule: geminin;PDBTitle: crystal structure of cdt1/geminin complex
78 c4a1eE_
not modelled
5.1
11
PDB header: ribosomeChain: E: PDB Molecule: 60s ribosomal protein l9;PDBTitle: t.thermophila 60s ribosomal subunit in complex with2 initiation factor 6. this file contains 5s rrna, 5.8s rrna3 and proteins of molecule 1
79 d1f6wa_
not modelled
5.0
5
Fold: alpha/beta-HydrolasesSuperfamily: alpha/beta-HydrolasesFamily: Acetylcholinesterase-like