| 1 | d1s0aa_
|
|
 |
100.0 |
98 |
Fold:PLP-dependent transferase-like
Superfamily:PLP-dependent transferases
Family:GABA-aminotransferase-like |
| 2 | c3fcrA_
|
|
 |
100.0 |
24 |
PDB header:transferase
Chain: A: PDB Molecule:putative aminotransferase;
PDBTitle: crystal structure of putative aminotransferase (yp_614685.1) from2 silicibacter sp. tm1040 at 1.80 a resolution
|
| 3 | c3lv2A_
|
|
 |
100.0 |
50 |
PDB header:transferase
Chain: A: PDB Molecule:adenosylmethionine-8-amino-7-oxononanoate aminotransferase;
PDBTitle: crystal structure of mycobacterium tuberculosis 7,8-diaminopelargonic2 acid synthase in complex with substrate analog sinefungin
|
| 4 | c3hmuA_
|
|
 |
100.0 |
28 |
PDB header:transferase
Chain: A: PDB Molecule:aminotransferase, class iii;
PDBTitle: crystal structure of a class iii aminotransferase from2 silicibacter pomeroyi
|
| 5 | c3n5mD_
|
|
 |
100.0 |
31 |
PDB header:transferase
Chain: D: PDB Molecule:adenosylmethionine-8-amino-7-oxononanoate aminotransferase;
PDBTitle: crystals structure of a bacillus anthracis aminotransferase
|
| 6 | c3i5tB_
|
|
 |
100.0 |
28 |
PDB header:transferase
Chain: B: PDB Molecule:aminotransferase;
PDBTitle: crystal structure of aminotransferase prk07036 from rhodobacter2 sphaeroides kd131
|
| 7 | c3bv0A_
|
|
 |
100.0 |
50 |
PDB header:transferase
Chain: A: PDB Molecule:adenosylmethionine-8-amino-7-oxononanoate aminotransferase;
PDBTitle: crystal structure of plp bound 7,8-diaminopelargonic acid synthase in2 mycobacterium tuberculosis
|
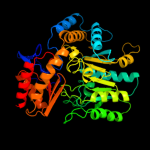
| 8 | c3a8uX_
|
|
 |
100.0 |
29 |
PDB header:transferase
Chain: X: PDB Molecule:omega-amino acid--pyruvate aminotransferase;
PDBTitle: crystal structure of omega-amino acid:pyruvate aminotransferase
|
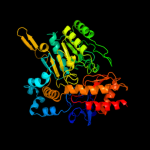
| 9 | d1ohwa_
|
|
 |
100.0 |
20 |
Fold:PLP-dependent transferase-like
Superfamily:PLP-dependent transferases
Family:GABA-aminotransferase-like |
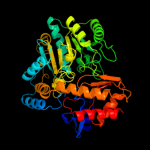
| 10 | c3oksB_
|
|
 |
100.0 |
27 |
PDB header:transferase
Chain: B: PDB Molecule:4-aminobutyrate transaminase;
PDBTitle: crystal structure of 4-aminobutyrate transaminase from mycobacterium2 smegmatis
|
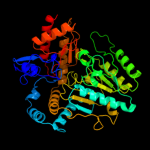
| 11 | c2cjdA_
|
|
 |
100.0 |
23 |
PDB header:transferase
Chain: A: PDB Molecule:l-lysine-epsilon aminotransferase;
PDBTitle: lysine aminotransferase from m. tuberculosis in external2 aldimine form
|
| 12 | c2eo5A_
|
|
 |
100.0 |
26 |
PDB header:transferase
Chain: A: PDB Molecule:419aa long hypothetical aminotransferase;
PDBTitle: crystal structure of 4-aminobutyrate aminotransferase from sulfolobus2 tokodaii strain7
|
| 13 | d1zoda1
|
|
 |
100.0 |
23 |
Fold:PLP-dependent transferase-like
Superfamily:PLP-dependent transferases
Family:GABA-aminotransferase-like |
| 14 | c3dodA_
|
|
 |
100.0 |
33 |
PDB header:transferase
Chain: A: PDB Molecule:adenosylmethionine-8-amino-7-oxononanoate aminotransferase;
PDBTitle: crystal structure of plp bound 7,8-diaminopelargonic acid synthase in2 bacillus subtilis
|
| 15 | c2e7uA_
|
|
 |
100.0 |
26 |
PDB header:isomerase
Chain: A: PDB Molecule:glutamate-1-semialdehyde 2,1-aminomutase;
PDBTitle: crystal structure of glutamate-1-semialdehyde 2,1-aminomutase from2 thermus thermophilus hb8
|
| 16 | c3nuiA_
|
|
 |
100.0 |
26 |
PDB header:transferase
Chain: A: PDB Molecule:pyruvate transaminase;
PDBTitle: crystal structure of omega-transferase from vibrio fluvialis js17
|
| 17 | c3bs8A_
|
|
 |
100.0 |
23 |
PDB header:isomerase
Chain: A: PDB Molecule:glutamate-1-semialdehyde 2,1-aminomutase;
PDBTitle: crystal structure of glutamate 1-semialdehyde aminotransferase2 complexed with pyridoxamine-5'-phosphate from bacillus subtilis
|
| 18 | d1sffa_
|
|
 |
100.0 |
28 |
Fold:PLP-dependent transferase-like
Superfamily:PLP-dependent transferases
Family:GABA-aminotransferase-like |
| 19 | d2gsaa_
|
|
 |
100.0 |
23 |
Fold:PLP-dependent transferase-like
Superfamily:PLP-dependent transferases
Family:GABA-aminotransferase-like |
| 20 | c3dxvA_
|
|
 |
100.0 |
28 |
PDB header:isomerase
Chain: A: PDB Molecule:alpha-amino-epsilon-caprolactam racemase;
PDBTitle: the crystal structure of alpha-amino-epsilon-caprolactam racemase from2 achromobacter obae
|
| 21 | d2byla1 |
|
not modelled |
100.0 |
27 |
Fold:PLP-dependent transferase-like
Superfamily:PLP-dependent transferases
Family:GABA-aminotransferase-like |
| 22 | c1oatB_ |
|
not modelled |
100.0 |
28 |
PDB header:aminotransferase
Chain: B: PDB Molecule:ornithine aminotransferase;
PDBTitle: ornithine aminotransferase
|
| 23 | c3ruyB_ |
|
not modelled |
100.0 |
27 |
PDB header:transferase
Chain: B: PDB Molecule:ornithine aminotransferase;
PDBTitle: crystal structure of the ornithine-oxo acid transaminase rocd from2 bacillus anthracis
|
| 24 | c2zsmA_ |
|
not modelled |
100.0 |
22 |
PDB header:isomerase
Chain: A: PDB Molecule:glutamate-1-semialdehyde 2,1-aminomutase;
PDBTitle: crystal structure of glutamate-1-semialdehyde 2,1-2 aminomutase from aeropyrum pernix, hexagonal form
|
| 25 | c2ordA_ |
|
not modelled |
100.0 |
28 |
PDB header:transferase
Chain: A: PDB Molecule:acetylornithine aminotransferase;
PDBTitle: crystal structure of acetylornithine aminotransferase (ec 2.6.1.11)2 (acoat) (tm1785) from thermotoga maritima at 1.40 a resolution
|
| 26 | c3l44A_ |
|
not modelled |
100.0 |
21 |
PDB header:isomerase
Chain: A: PDB Molecule:glutamate-1-semialdehyde 2,1-aminomutase 1;
PDBTitle: crystal structure of bacillus anthracis heml-1, glutamate semialdehyde2 aminotransferase
|
| 27 | d1z7da1 |
|
not modelled |
100.0 |
27 |
Fold:PLP-dependent transferase-like
Superfamily:PLP-dependent transferases
Family:GABA-aminotransferase-like |
| 28 | d1vefa1 |
|
not modelled |
100.0 |
32 |
Fold:PLP-dependent transferase-like
Superfamily:PLP-dependent transferases
Family:GABA-aminotransferase-like |
| 29 | c1z7dE_ |
|
not modelled |
100.0 |
27 |
PDB header:transferase
Chain: E: PDB Molecule:ornithine aminotransferase;
PDBTitle: ornithine aminotransferase py00104 from plasmodium yoelii
|
| 30 | c2pb2B_ |
|
not modelled |
100.0 |
30 |
PDB header:transferase
Chain: B: PDB Molecule:acetylornithine/succinyldiaminopimelate aminotransferase;
PDBTitle: structure of biosynthetic n-acetylornithine aminotransferase from2 salmonella typhimurium: studies on substrate specificity and3 inhibitor binding
|
| 31 | c3nx3A_ |
|
not modelled |
100.0 |
28 |
PDB header:transferase
Chain: A: PDB Molecule:acetylornithine aminotransferase;
PDBTitle: crystal structure of acetylornithine aminotransferase (argd) from2 campylobacter jejuni
|
| 32 | c3i4jC_ |
|
not modelled |
100.0 |
28 |
PDB header:transferase
Chain: C: PDB Molecule:aminotransferase, class iii;
PDBTitle: crystal structure of aminotransferase, class iii from2 deinococcus radiodurans
|
| 33 | c2cy8A_ |
|
not modelled |
100.0 |
23 |
PDB header:transferase
Chain: A: PDB Molecule:d-phenylglycine aminotransferase;
PDBTitle: crystal structure of d-phenylglycine aminotransferase (d-phgat) from2 pseudomonas strutzeri st-201
|
| 34 | c2eh6A_ |
|
not modelled |
100.0 |
30 |
PDB header:transferase
Chain: A: PDB Molecule:acetylornithine aminotransferase;
PDBTitle: crystal structure of acetylornithine aminotransferase from aquifex2 aeolicus vf5
|
| 35 | d2cfba1 |
|
not modelled |
100.0 |
27 |
Fold:PLP-dependent transferase-like
Superfamily:PLP-dependent transferases
Family:GABA-aminotransferase-like |
| 36 | c2w8wA_ |
|
not modelled |
100.0 |
13 |
PDB header:transferase
Chain: A: PDB Molecule:serine palmitoyltransferase;
PDBTitle: n100y spt with plp-ser
|
| 37 | c3a2bA_ |
|
not modelled |
100.0 |
14 |
PDB header:transferase
Chain: A: PDB Molecule:serine palmitoyltransferase;
PDBTitle: crystal structure of serine palmitoyltransferase from sphingobacterium2 multivorum with substrate l-serine
|
| 38 | d1fc4a_ |
|
not modelled |
100.0 |
18 |
Fold:PLP-dependent transferase-like
Superfamily:PLP-dependent transferases
Family:GABA-aminotransferase-like |
| 39 | d2bwna1 |
|
not modelled |
100.0 |
16 |
Fold:PLP-dependent transferase-like
Superfamily:PLP-dependent transferases
Family:GABA-aminotransferase-like |
| 40 | d1bs0a_ |
|
not modelled |
100.0 |
19 |
Fold:PLP-dependent transferase-like
Superfamily:PLP-dependent transferases
Family:GABA-aminotransferase-like |
| 41 | c3tqxA_ |
|
not modelled |
100.0 |
14 |
PDB header:transferase
Chain: A: PDB Molecule:2-amino-3-ketobutyrate coenzyme a ligase;
PDBTitle: structure of the 2-amino-3-ketobutyrate coenzyme a ligase (kbl) from2 coxiella burnetii
|
| 42 | c3hqtB_ |
|
not modelled |
100.0 |
15 |
PDB header:transferase
Chain: B: PDB Molecule:cai-1 autoinducer synthase;
PDBTitle: plp-dependent acyl-coa transferase cqsa
|
| 43 | c3pj0D_ |
|
not modelled |
100.0 |
13 |
PDB header:lyase
Chain: D: PDB Molecule:lmo0305 protein;
PDBTitle: crystal structure of a putative l-allo-threonine aldolase (lmo0305)2 from listeria monocytogenes egd-e at 1.80 a resolution
|
| 44 | c3ke3A_ |
|
not modelled |
100.0 |
15 |
PDB header:transferase
Chain: A: PDB Molecule:putative serine-pyruvate aminotransferase;
PDBTitle: crystal structure of putative serine-pyruvate aminotransferase2 (yp_263484.1) from psychrobacter arcticum 273-4 at 2.20 a resolution
|
| 45 | d1m6sa_ |
|
not modelled |
100.0 |
16 |
Fold:PLP-dependent transferase-like
Superfamily:PLP-dependent transferases
Family:AAT-like |
| 46 | d1c7ga_ |
|
not modelled |
100.0 |
13 |
Fold:PLP-dependent transferase-like
Superfamily:PLP-dependent transferases
Family:Beta-eliminating lyases |
| 47 | d1qz9a_ |
|
not modelled |
100.0 |
16 |
Fold:PLP-dependent transferase-like
Superfamily:PLP-dependent transferases
Family:Cystathionine synthase-like |
| 48 | d1svva_ |
|
not modelled |
100.0 |
13 |
Fold:PLP-dependent transferase-like
Superfamily:PLP-dependent transferases
Family:AAT-like |
| 49 | d1wyub1 |
|
not modelled |
100.0 |
17 |
Fold:PLP-dependent transferase-like
Superfamily:PLP-dependent transferases
Family:Glycine dehydrogenase subunits (GDC-P) |
| 50 | d1tpla_ |
|
not modelled |
100.0 |
13 |
Fold:PLP-dependent transferase-like
Superfamily:PLP-dependent transferases
Family:Beta-eliminating lyases |
| 51 | d1wsta1 |
|
not modelled |
100.0 |
11 |
Fold:PLP-dependent transferase-like
Superfamily:PLP-dependent transferases
Family:AAT-like |
| 52 | c3lwsF_ |
|
not modelled |
100.0 |
15 |
PDB header:lyase
Chain: F: PDB Molecule:aromatic amino acid beta-eliminating
PDBTitle: crystal structure of putative aromatic amino acid beta-2 eliminating lyase/threonine aldolase. (yp_001813866.1) from3 exiguobacterium sp. 255-15 at 2.00 a resolution
|
| 53 | d1v72a1 |
|
not modelled |
100.0 |
15 |
Fold:PLP-dependent transferase-like
Superfamily:PLP-dependent transferases
Family:AAT-like |
| 54 | d1ax4a_ |
|
not modelled |
100.0 |
14 |
Fold:PLP-dependent transferase-like
Superfamily:PLP-dependent transferases
Family:Beta-eliminating lyases |
| 55 | c3mafB_ |
|
not modelled |
100.0 |
14 |
PDB header:lyase
Chain: B: PDB Molecule:sphingosine-1-phosphate lyase;
PDBTitle: crystal structure of stspl (asymmetric form)
|
| 56 | c3f9tB_ |
|
not modelled |
100.0 |
12 |
PDB header:lyase
Chain: B: PDB Molecule:l-tyrosine decarboxylase mfna;
PDBTitle: crystal structure of l-tyrosine decarboxylase mfna (ec 4.1.1.25)2 (np_247014.1) from methanococcus jannaschii at 2.11 a resolution
|
| 57 | d1x0ma1 |
|
not modelled |
100.0 |
8 |
Fold:PLP-dependent transferase-like
Superfamily:PLP-dependent transferases
Family:AAT-like |
| 58 | c2hzpA_ |
|
not modelled |
100.0 |
12 |
PDB header:hydrolase
Chain: A: PDB Molecule:kynureninase;
PDBTitle: crystal structure of homo sapiens kynureninase
|
| 59 | d2v1pa1 |
|
not modelled |
100.0 |
12 |
Fold:PLP-dependent transferase-like
Superfamily:PLP-dependent transferases
Family:Beta-eliminating lyases |
| 60 | c3e9kA_ |
|
not modelled |
100.0 |
12 |
PDB header:hydrolase
Chain: A: PDB Molecule:kynureninase;
PDBTitle: crystal structure of homo sapiens kynureninase-3-hydroxyhippuric acid2 inhibitor complex
|
| 61 | d1lc5a_ |
|
not modelled |
99.9 |
15 |
Fold:PLP-dependent transferase-like
Superfamily:PLP-dependent transferases
Family:AAT-like |
| 62 | d1wyua1 |
|
not modelled |
99.9 |
13 |
Fold:PLP-dependent transferase-like
Superfamily:PLP-dependent transferases
Family:Glycine dehydrogenase subunits (GDC-P) |
| 63 | c2ogeC_ |
|
not modelled |
99.9 |
13 |
PDB header:transferase
Chain: C: PDB Molecule:transaminase;
PDBTitle: x-ray structure of s. venezuelae desv in its internal2 aldimine form
|
| 64 | c2hdyA_ |
|
not modelled |
99.9 |
12 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:selenocysteine lyase;
PDBTitle: structure of human selenocysteine lyase
|
| 65 | d1js3a_ |
|
not modelled |
99.9 |
11 |
Fold:PLP-dependent transferase-like
Superfamily:PLP-dependent transferases
Family:Pyridoxal-dependent decarboxylase |
| 66 | d1jf9a_ |
|
not modelled |
99.9 |
12 |
Fold:PLP-dependent transferase-like
Superfamily:PLP-dependent transferases
Family:Cystathionine synthase-like |
| 67 | d1c7na_ |
|
not modelled |
99.9 |
12 |
Fold:PLP-dependent transferase-like
Superfamily:PLP-dependent transferases
Family:Cystathionine synthase-like |
| 68 | d1xi9a_ |
|
not modelled |
99.9 |
11 |
Fold:PLP-dependent transferase-like
Superfamily:PLP-dependent transferases
Family:AAT-like |
| 69 | d1o4sa_ |
|
not modelled |
99.9 |
11 |
Fold:PLP-dependent transferase-like
Superfamily:PLP-dependent transferases
Family:AAT-like |
| 70 | d1eg5a_ |
|
not modelled |
99.9 |
11 |
Fold:PLP-dependent transferase-like
Superfamily:PLP-dependent transferases
Family:Cystathionine synthase-like |
| 71 | c2zc0C_ |
|
not modelled |
99.9 |
11 |
PDB header:transferase
Chain: C: PDB Molecule:alanine glyoxylate transaminase;
PDBTitle: crystal structure of an archaeal alanine:glyoxylate aminotransferase
|
| 72 | c2x5dD_ |
|
not modelled |
99.9 |
9 |
PDB header:transferase
Chain: D: PDB Molecule:probable aminotransferase;
PDBTitle: crystal structure of a probable aminotransferase from2 pseudomonas aeruginosa
|
| 73 | c2dkjB_ |
|
not modelled |
99.9 |
14 |
PDB header:transferase
Chain: B: PDB Molecule:serine hydroxymethyltransferase;
PDBTitle: crystal structure of t.th.hb8 serine hydroxymethyltransferase
|
| 74 | c3ftbA_ |
|
not modelled |
99.9 |
13 |
PDB header:transferase
Chain: A: PDB Molecule:histidinol-phosphate aminotransferase;
PDBTitle: the crystal structure of the histidinol-phosphate2 aminotransferase from clostridium acetobutylicum
|
| 75 | d1elua_ |
|
not modelled |
99.9 |
11 |
Fold:PLP-dependent transferase-like
Superfamily:PLP-dependent transferases
Family:Cystathionine synthase-like |
| 76 | d1b5pa_ |
|
not modelled |
99.9 |
13 |
Fold:PLP-dependent transferase-like
Superfamily:PLP-dependent transferases
Family:AAT-like |
| 77 | d1kl1a_ |
|
not modelled |
99.9 |
14 |
Fold:PLP-dependent transferase-like
Superfamily:PLP-dependent transferases
Family:GABA-aminotransferase-like |
| 78 | c2dr1A_ |
|
not modelled |
99.9 |
10 |
PDB header:transferase
Chain: A: PDB Molecule:386aa long hypothetical serine aminotransferase;
PDBTitle: crystal structure of the ph1308 protein from pyrococcus horikoshii ot3
|
| 79 | c3cbfA_ |
|
not modelled |
99.9 |
11 |
PDB header:transferase
Chain: A: PDB Molecule:alpha-aminodipate aminotransferase;
PDBTitle: crystal structure of lysn, alpha-aminoadipate2 aminotransferase, from thermus thermophilus hb27
|
| 80 | c2douA_ |
|
not modelled |
99.9 |
14 |
PDB header:transferase
Chain: A: PDB Molecule:probable n-succinyldiaminopimelate aminotransferase;
PDBTitle: probable n-succinyldiaminopimelate aminotransferase (ttha0342) from2 thermus thermophilus hb8
|
| 81 | c3kaxB_ |
|
not modelled |
99.9 |
10 |
PDB header:lyase
Chain: B: PDB Molecule:aminotransferase, classes i and ii;
PDBTitle: crystal structure of a putative c-s lyase from bacillus anthracis
|
| 82 | d1j32a_ |
|
not modelled |
99.9 |
12 |
Fold:PLP-dependent transferase-like
Superfamily:PLP-dependent transferases
Family:AAT-like |
| 83 | c3l8aB_ |
|
not modelled |
99.9 |
11 |
PDB header:lyase
Chain: B: PDB Molecule:putative aminotransferase, probable beta-cystathionase;
PDBTitle: crystal structure of metc from streptococcus mutans
|
| 84 | c3getA_ |
|
not modelled |
99.9 |
13 |
PDB header:transferase
Chain: A: PDB Molecule:histidinol-phosphate aminotransferase;
PDBTitle: crystal structure of putative histidinol-phosphate aminotransferase2 (np_281508.1) from campylobacter jejuni at 2.01 a resolution
|
| 85 | c3qguB_ |
|
not modelled |
99.9 |
7 |
PDB header:transferase
Chain: B: PDB Molecule:ll-diaminopimelate aminotransferase;
PDBTitle: l,l-diaminopimelate aminotransferase from chalmydomonas reinhardtii
|
| 86 | c3caiA_ |
|
not modelled |
99.9 |
14 |
PDB header:transferase
Chain: A: PDB Molecule:possible aminotransferase;
PDBTitle: crystal structure of mycobacterium tuberculosis rv3778c2 protein
|
| 87 | d1t3ia_ |
|
not modelled |
99.9 |
11 |
Fold:PLP-dependent transferase-like
Superfamily:PLP-dependent transferases
Family:Cystathionine synthase-like |
| 88 | c3jtxB_ |
|
not modelled |
99.9 |
9 |
PDB header:transferase
Chain: B: PDB Molecule:aminotransferase;
PDBTitle: crystal structure of aminotransferase (np_283882.1) from neisseria2 meningitidis z2491 at 1.91 a resolution
|
| 89 | d1vp4a_ |
|
not modelled |
99.9 |
11 |
Fold:PLP-dependent transferase-like
Superfamily:PLP-dependent transferases
Family:AAT-like |
| 90 | c3fdbA_ |
|
not modelled |
99.9 |
11 |
PDB header:transferase
Chain: A: PDB Molecule:putative plp-dependent beta-cystathionase;
PDBTitle: crystal structure of a putative plp-dependent beta-cystathionase2 (aecd, dip1736) from corynebacterium diphtheriae at 1.99 a resolution
|
| 91 | c3op7A_ |
|
not modelled |
99.9 |
10 |
PDB header:transferase
Chain: A: PDB Molecule:aminotransferase class i and ii;
PDBTitle: crystal structure of a plp-dependent aminotransferase (zp_03625122.1)2 from streptococcus suis 89-1591 at 1.70 a resolution
|
| 92 | c3lvmB_ |
|
not modelled |
99.9 |
13 |
PDB header:transferase
Chain: B: PDB Molecule:cysteine desulfurase;
PDBTitle: crystal structure of e.coli iscs
|
| 93 | c3eibB_ |
|
not modelled |
99.9 |
8 |
PDB header:transferase
Chain: B: PDB Molecule:ll-diaminopimelate aminotransferase;
PDBTitle: crystal structure of k270n variant of ll-diaminopimelate2 aminotransferase from arabidopsis thaliana
|
| 94 | c3dc1A_ |
|
not modelled |
99.9 |
9 |
PDB header:transferase
Chain: A: PDB Molecule:kynurenine/alpha-aminoadipate aminotransferase
PDBTitle: crystal structure of kynurenine aminotransferase ii complex with2 alpha-ketoglutarate
|
| 95 | c3k40B_ |
|
not modelled |
99.9 |
11 |
PDB header:lyase
Chain: B: PDB Molecule:aromatic-l-amino-acid decarboxylase;
PDBTitle: crystal structure of drosophila 3,4-dihydroxyphenylalanine2 decarboxylase
|
| 96 | c3nysA_ |
|
not modelled |
99.9 |
14 |
PDB header:transferase
Chain: A: PDB Molecule:aminotransferase wbpe;
PDBTitle: x-ray structure of the k185a mutant of wbpe (wlbe) from pseudomonas2 aeruginosa in complex with plp at 1.45 angstrom resolution
|
| 97 | c3hdoB_ |
|
not modelled |
99.9 |
11 |
PDB header:transferase
Chain: B: PDB Molecule:histidinol-phosphate aminotransferase;
PDBTitle: crystal structure of a histidinol-phosphate aminotransferase from2 geobacter metallireducens
|
| 98 | c2okkA_ |
|
not modelled |
99.9 |
11 |
PDB header:lyase
Chain: A: PDB Molecule:glutamate decarboxylase 2;
PDBTitle: the x-ray crystal structure of the 65kda isoform of glutamic acid2 decarboxylase (gad65)
|
| 99 | c3fkdC_ |
|
not modelled |
99.9 |
12 |
PDB header:lyase
Chain: C: PDB Molecule:l-threonine-o-3-phosphate decarboxylase;
PDBTitle: the crystal structure of l-threonine-o-3-phosphate2 decarboxylase from porphyromonas gingivalis
|
| 100 | d1d2fa_ |
|
not modelled |
99.9 |
12 |
Fold:PLP-dependent transferase-like
Superfamily:PLP-dependent transferases
Family:Cystathionine synthase-like |
| 101 | c3pdxA_ |
|
not modelled |
99.9 |
13 |
PDB header:transferase
Chain: A: PDB Molecule:tyrosine aminotransferase;
PDBTitle: crystal structural of mouse tyrosine aminotransferase
|
| 102 | c3dr4B_ |
|
not modelled |
99.9 |
14 |
PDB header:transferase
Chain: B: PDB Molecule:putative perosamine synthetase;
PDBTitle: gdp-perosamine synthase k186a mutant from caulobacter2 crescentus with bound sugar ligand
|
| 103 | c2jisA_ |
|
not modelled |
99.9 |
14 |
PDB header:lyase
Chain: A: PDB Molecule:cysteine sulfinic acid decarboxylase;
PDBTitle: human cysteine sulfinic acid decarboxylase (csad) in2 complex with plp.
|
| 104 | c3uwcA_ |
|
not modelled |
99.9 |
12 |
PDB header:transferase
Chain: A: PDB Molecule:nucleotide-sugar aminotransferase;
PDBTitle: structure of an aminotransferase (degt-dnrj-eryc1-strs family) from2 coxiella burnetii in complex with pmp
|
| 105 | d1v2da_ |
|
not modelled |
99.9 |
14 |
Fold:PLP-dependent transferase-like
Superfamily:PLP-dependent transferases
Family:AAT-like |
| 106 | c1d2fB_ |
|
not modelled |
99.9 |
12 |
PDB header:transferase
Chain: B: PDB Molecule:maly protein;
PDBTitle: x-ray structure of maly from escherichia coli: a pyridoxal-5'-2 phosphate-dependent enzyme acting as a modulator in mal gene3 expression
|
| 107 | d1o69a_ |
|
not modelled |
99.9 |
13 |
Fold:PLP-dependent transferase-like
Superfamily:PLP-dependent transferases
Family:GABA-aminotransferase-like |
| 108 | d2fnua1 |
|
not modelled |
99.9 |
9 |
Fold:PLP-dependent transferase-like
Superfamily:PLP-dependent transferases
Family:GABA-aminotransferase-like |
| 109 | d1mdoa_ |
|
not modelled |
99.9 |
16 |
Fold:PLP-dependent transferase-like
Superfamily:PLP-dependent transferases
Family:GABA-aminotransferase-like |
| 110 | c3frkB_ |
|
not modelled |
99.9 |
12 |
PDB header:transferase
Chain: B: PDB Molecule:qdtb;
PDBTitle: x-ray structure of qdtb from t. thermosaccharolyticum in2 complex with a plp:tdp-3-aminoquinovose aldimine
|
| 111 | c3dzzB_ |
|
not modelled |
99.9 |
11 |
PDB header:transferase
Chain: B: PDB Molecule:putative pyridoxal 5'-phosphate-dependent c-s lyase;
PDBTitle: crystal structure of a putative plp-dependent aminotransferase2 (lbul_1103) from lactobacillus delbrueckii subsp. at 1.61 a3 resolution
|
| 112 | d1b9ha_ |
|
not modelled |
99.9 |
12 |
Fold:PLP-dependent transferase-like
Superfamily:PLP-dependent transferases
Family:GABA-aminotransferase-like |
| 113 | d1p3wa_ |
|
not modelled |
99.9 |
13 |
Fold:PLP-dependent transferase-like
Superfamily:PLP-dependent transferases
Family:Cystathionine synthase-like |
| 114 | c3mc6C_ |
|
not modelled |
99.9 |
10 |
PDB header:lyase
Chain: C: PDB Molecule:sphingosine-1-phosphate lyase;
PDBTitle: crystal structure of scdpl1
|
| 115 | d1vjoa_ |
|
not modelled |
99.9 |
14 |
Fold:PLP-dependent transferase-like
Superfamily:PLP-dependent transferases
Family:Cystathionine synthase-like |
| 116 | c3nraA_ |
|
not modelled |
99.9 |
15 |
PDB header:transferase
Chain: A: PDB Molecule:aspartate aminotransferase;
PDBTitle: crystal structure of an aspartate aminotransferase (yp_354942.1) from2 rhodobacter sphaeroides 2.4.1 at 2.15 a resolution
|
| 117 | d2f8ja1 |
|
not modelled |
99.9 |
13 |
Fold:PLP-dependent transferase-like
Superfamily:PLP-dependent transferases
Family:AAT-like |
| 118 | c3ly1C_ |
|
not modelled |
99.9 |
13 |
PDB header:transferase
Chain: C: PDB Molecule:putative histidinol-phosphate aminotransferase;
PDBTitle: crystal structure of putative histidinol-phosphate aminotransferase2 (yp_050345.1) from erwinia carotovora atroseptica scri1043 at 1.80 a3 resolution
|
| 119 | c2r0tA_ |
|
not modelled |
99.9 |
14 |
PDB header:transferase
Chain: A: PDB Molecule:pyridoxamine 5-phosphate-dependent dehydrase;
PDBTitle: crystal sructure of gdp-4-keto-6-deoxymannose-3-dehydratase2 with a trapped plp-glutamate geminal diamine
|
| 120 | c3cq6E_ |
|
not modelled |
99.9 |
14 |
PDB header:transferase
Chain: E: PDB Molecule:histidinol-phosphate aminotransferase;
PDBTitle: histidinol-phosphate aminotransferase from corynebacterium2 glutamicum holo-form (plp covalently bound )
|