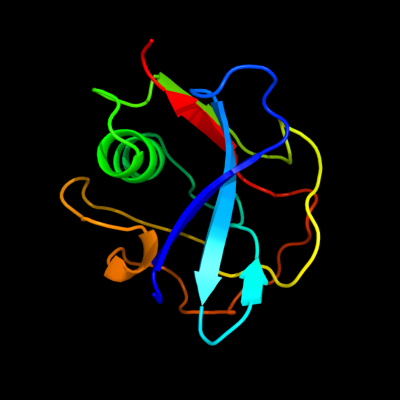
1 d1by2a_
100.0
49
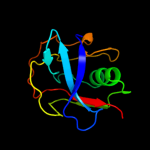
Fold: SRCR-likeSuperfamily: SRCR-likeFamily: Scavenger receptor cysteine-rich (SRCR) domain2 c2oyaA_
100.0
46
PDB header: ligand binding proteinChain: A: PDB Molecule: macrophage receptor marco;PDBTitle: crystal structure analysis of the dimeric form of the srcr domain of2 mouse marco
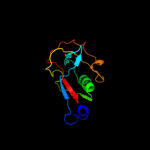
3 c2jopA_
100.0
28
PDB header: immune systemChain: A: PDB Molecule: t-cell surface glycoprotein cd5;PDBTitle: solution structure of the n-terminal extracellular domain2 of the lymphocyte receptor cd5 (cd5 domain 1)
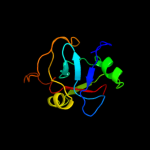
4 c2ottY_
100.0
33
PDB header: immune systemChain: Y: PDB Molecule: t-cell surface glycoprotein cd5;PDBTitle: crystal structure of cd5_diii
5 d1z8ga2
99.9
18
Fold: SRCR-likeSuperfamily: SRCR-likeFamily: Hepsin, N-terminal domain6 c1z8gA_
99.8
25
PDB header: hydrolase/hydrolase inhibitorChain: A: PDB Molecule: serine protease hepsin;PDBTitle: crystal structure of the extracellular region of the transmembrane2 serine protease hepsin with covalently bound preferred substrate.
7 c2xrcD_
96.8
24
PDB header: immune systemChain: D: PDB Molecule: human complement factor i;PDBTitle: human complement factor i
8 c2dvkA_
19.7
27
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: upf0130 protein ape0816;PDBTitle: crystal structure of hypothetical protein from aeropyrum pernix
9 d1tlja_
17.0
29
Fold: SSo0622-likeSuperfamily: SSo0622-likeFamily: SSo0622-like10 c1tljA_
17.0
29
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: hypothetical upf0130 protein sso0622;PDBTitle: crystal structure of conserved protein of unknown function2 sso0622 from sulfolobus solfataricus
11 c2lw6A_
15.9
27
PDB header: apoptosisChain: A: PDB Molecule: avrpiz-t protein;PDBTitle: solution structure of an avirulence protein avrpiz-t from pathogen2 magnaportheoryzae
12 d1pbya5
15.3
22
Fold: Streptavidin-likeSuperfamily: Quinohemoprotein amine dehydrogenase A chain, domain 3Family: Quinohemoprotein amine dehydrogenase A chain, domain 313 c2aklA_
13.2
24
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: phna-like protein pa0128;PDBTitle: solution structure for phn-a like protein pa0128 from2 pseudomonas aeruginosa
14 d2akka1
13.1
24
Fold: SH3-like barrelSuperfamily: Prokaryotic SH3-related domainFamily: PhnA-like15 d2akla1
12.7
24
Fold: SH3-like barrelSuperfamily: Prokaryotic SH3-related domainFamily: PhnA-like16 c2qg3B_
9.1
25
PDB header: unknown functionChain: B: PDB Molecule: upf0130 protein af_2059;PDBTitle: crystal structure of a tyw3 methyltransferase-like protein (af_2059)2 from archaeoglobus fulgidus dsm 4304 at 1.95 a resolution
17 c1iicA_
8.9
23
PDB header: transferaseChain: A: PDB Molecule: peptide n-myristoyltransferase;PDBTitle: crystal structure of saccharomyces cerevisiae n-myristoyltransferase2 with bound myristoylcoa
18 d2vxfa1
8.8
21
Fold: Sm-like foldSuperfamily: Sm-like ribonucleoproteinsFamily: LSM14 N-terminal domain-like19 c2p6fA_
8.7
23
PDB header: transferaseChain: A: PDB Molecule: glycylpeptide n-tetradecanoyltransferase;PDBTitle: crystal structures of saccharomyces cerevisiae n-myristoyltransferase2 with bound myristoyl-coa and inhibitors
20 c1dzlA_
8.6
16
PDB header: virusChain: A: PDB Molecule: late major capsid protein l1;PDBTitle: l1 protein of human papillomavirus 16
21 d1dzla_
not modelled
8.6
16
Fold: Nucleoplasmin-like/VP (viral coat and capsid proteins)Superfamily: Group I dsDNA virusesFamily: Papovaviridae-like VP22 d2ns0a1
not modelled
8.4
36
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: RHA1 ro06458-like23 c1jpeA_
not modelled
8.4
15
PDB header: electron transportChain: A: PDB Molecule: dsbd-alpha;PDBTitle: crystal structure of dsbd-alpha; the n-terminal domain of2 dsbd
24 c2vxeA_
not modelled
8.1
26
PDB header: transcriptionChain: A: PDB Molecule: cg10686-pa;PDBTitle: solution structure of the lsm domain of drosophila2 melanogaster tral (trailer hitch)
25 c2it3B_
not modelled
8.0
22
PDB header: structural genomics, unknown functionChain: B: PDB Molecule: upf0130 protein ph1069;PDBTitle: structure of ph1069 protein from pyrococcus horikoshii
26 d1tn3a_
not modelled
8.0
25
Fold: C-type lectin-likeSuperfamily: C-type lectin-likeFamily: C-type lectin domain27 d1iica2
not modelled
7.8
23
Fold: Acyl-CoA N-acyltransferases (Nat)Superfamily: Acyl-CoA N-acyltransferases (Nat)Family: N-myristoyl transferase, NMT28 c1iylC_
not modelled
7.8
28
PDB header: transferaseChain: C: PDB Molecule: myristoyl-coa:protein n-myristoyltransferase;PDBTitle: crystal structure of candida albicans n-myristoyltransferase with non-2 peptidic inhibitor
29 c1fwoA_
not modelled
7.7
19
PDB header: hydrolaseChain: A: PDB Molecule: oryzain beta chain;PDBTitle: the solution structure of a 35-residue fragment from the2 granulin/epithelin-like subdomain of rice oryzain beta3 (rob 382-416 (c398s,c399s,c407s,c413s))
30 d1oz7a_
not modelled
7.7
25
Fold: C-type lectin-likeSuperfamily: C-type lectin-likeFamily: C-type lectin domain31 c4dx94_
not modelled
7.6
23
PDB header: protein bindingChain: 4: PDB Molecule: integrin beta-1-binding protein 1;PDBTitle: icap1 in complex with integrin beta 1 cytoplasmic tail
32 c2r5kE_
not modelled
7.6
13
PDB header: viral proteinChain: E: PDB Molecule: major capsid protein l1;PDBTitle: pentamer structure of major capsid protein l1 of human2 papilloma virus type 11
33 c2k0rA_
not modelled
7.4
33
PDB header: oxidoreductaseChain: A: PDB Molecule: thiol:disulfide interchange protein dsbd;PDBTitle: solution structure of the c103s mutant of the n-terminal2 domain of dsbd from neisseria meningitidis
34 d1rxta2
not modelled
7.2
37
Fold: Acyl-CoA N-acyltransferases (Nat)Superfamily: Acyl-CoA N-acyltransferases (Nat)Family: N-myristoyl transferase, NMT35 d1l6pa_
not modelled
7.2
17
Fold: Immunoglobulin-like beta-sandwichSuperfamily: Thiol:disulfide interchange protein DsbD, N-terminal domain (DsbD-alpha)Family: Thiol:disulfide interchange protein DsbD, N-terminal domain (DsbD-alpha)36 d1iyka2
not modelled
7.1
30
Fold: Acyl-CoA N-acyltransferases (Nat)Superfamily: Acyl-CoA N-acyltransferases (Nat)Family: N-myristoyl transferase, NMT37 c3iu2B_
not modelled
6.9
29
PDB header: transferaseChain: B: PDB Molecule: glycylpeptide n-tetradecanoyltransferase 1;PDBTitle: crystal structure of human type-i n-myristoyltransferase with bound2 myristoyl-coa and inhibitor ddd90096
38 c1nj3A_
not modelled
6.6
26
PDB header: protein bindingChain: A: PDB Molecule: npl4;PDBTitle: structure and ubiquitin interactions of the conserved nzf2 domain of npl4
39 c2a45J_
not modelled
6.4
32
PDB header: hydrolase/hydrolase inhibitorChain: J: PDB Molecule: fibrinogen alpha chain;PDBTitle: crystal structure of the complex between thrombin and the central "e"2 region of fibrin
40 c3a57A_
not modelled
6.3
32
PDB header: toxinChain: A: PDB Molecule: thermostable direct hemolysin 2;PDBTitle: crystal structure of thermostable direct hemolysin
41 d1wk1a_
not modelled
6.3
32
Fold: C-type lectin-likeSuperfamily: C-type lectin-likeFamily: C-type lectin domain42 c2vpnB_
not modelled
6.3
8
PDB header: transportChain: B: PDB Molecule: periplasmic substrate binding protein;PDBTitle: high-resolution structure of the periplasmic ectoine-2 binding protein from teaabc trap-transporter of halomonas3 elongata
43 c3ke2A_
not modelled
6.2
26
PDB header: unknown functionChain: A: PDB Molecule: uncharacterized protein yp_928783.1;PDBTitle: crystal structure of a duf2131 family protein (sama_2911) from2 shewanella amazonensis sb2b at 2.50 a resolution
44 c2jtkA_
not modelled
6.0
31
PDB header: signaling proteinChain: A: PDB Molecule: dickkopf-related protein 2;PDBTitle: a functional domain of a wnt signal protein
45 d1jmxa5
not modelled
5.9
13
Fold: Streptavidin-likeSuperfamily: Quinohemoprotein amine dehydrogenase A chain, domain 3Family: Quinohemoprotein amine dehydrogenase A chain, domain 346 c4g5eD_
not modelled
5.9
22
PDB header: oxidoreductaseChain: D: PDB Molecule: 2,4,6-trichlorophenol 4-monooxygenase;PDBTitle: 2,4,6-trichlorophenol 4-monooxygenase
47 c3fxbB_
not modelled
5.7
15
PDB header: transport proteinChain: B: PDB Molecule: trap dicarboxylate transporter, dctp subunit;PDBTitle: crystal structure of the ectoine-binding protein ueha
48 d2pf5a1
not modelled
5.6
27
Fold: C-type lectin-likeSuperfamily: C-type lectin-likeFamily: Link domain49 d1k42a_
not modelled
5.5
15
Fold: Galactose-binding domain-likeSuperfamily: Galactose-binding domain-likeFamily: CBM4/950 c3djmA_
not modelled
5.4
24
PDB header: unknown functionChain: A: PDB Molecule: uncharacterized protein duf427;PDBTitle: crystal structure of a protein of unknown function from duf427 family2 (rsph17029_0682) from rhodobacter sphaeroides 2.4.1 at 2.51 a3 resolution
51 c3iyjE_
not modelled
5.4
9
PDB header: virusChain: E: PDB Molecule: major capsid protein l1;PDBTitle: bovine papillomavirus type 1 outer capsid
52 c4ag4A_
not modelled
5.4
13
PDB header: immune system/transferaseChain: A: PDB Molecule: epithelial discoidin domain-containing receptor 1;PDBTitle: crystal structure of a ddr1-fab complex