| 1 | c2k4zA_
|
|
 |
100.0 |
15 |
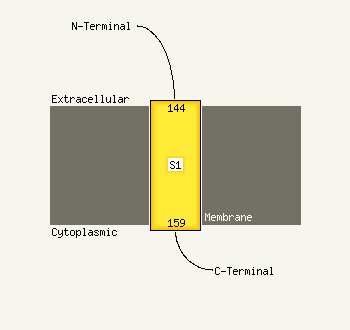
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:dsrr;
PDBTitle: solution nmr structure of allochromatium vinosum dsrr:2 northeast structural genomics consortium target op5
|
| 2 | d1veha_
|
|
 |
100.0 |
29 |
Fold:Alpha-lytic protease prodomain-like
Superfamily:Fe-S cluster assembly (FSCA) domain-like
Family:NifU C-terminal domain-like |
| 3 | d1xhja_
|
|
 |
100.0 |
31 |
Fold:Alpha-lytic protease prodomain-like
Superfamily:Fe-S cluster assembly (FSCA) domain-like
Family:NifU C-terminal domain-like |
| 4 | d1s98a_
|
|
 |
100.0 |
26 |
Fold:HesB-like domain
Superfamily:HesB-like domain
Family:HesB-like domain |
| 5 | c2d2aA_
|
|
 |
99.9 |
21 |
PDB header:metal transport
Chain: A: PDB Molecule:sufa protein;
PDBTitle: crystal structure of escherichia coli sufa involved in2 biosynthesis of iron-sulfur clusters
|
| 6 | d1nwba_
|
|
 |
99.9 |
26 |
Fold:HesB-like domain
Superfamily:HesB-like domain
Family:HesB-like domain |
| 7 | c1x0gA_
|
|
 |
99.9 |
20 |
PDB header:metal binding protein
Chain: A: PDB Molecule:isca;
PDBTitle: crystal structure of isca with the [2fe-2s] cluster
|
| 8 | c2jnvA_
|
|
 |
99.9 |
34 |
PDB header:metal transport
Chain: A: PDB Molecule:nifu-like protein 1, chloroplast;
PDBTitle: solution structure of c-terminal domain of nifu-like2 protein from oryza sativa
|
| 9 | c2apnA_
|
|
 |
99.9 |
23 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:protein hi1723;
PDBTitle: hi1723 solution structure
|
| 10 | c2z51A_
|
|
 |
99.9 |
35 |
PDB header:metal transport
Chain: A: PDB Molecule:nifu-like protein 2, chloroplast;
PDBTitle: crystal structure of arabidopsis cnfu involved in iron-2 sulfur cluster biosynthesis
|
| 11 | d1th5a1
|
|
 |
99.8 |
25 |
Fold:Alpha-lytic protease prodomain-like
Superfamily:Fe-S cluster assembly (FSCA) domain-like
Family:NifU C-terminal domain-like |
| 12 | d2p2ea1
|
|
 |
97.7 |
16 |
Fold:HesB-like domain
Superfamily:HesB-like domain
Family:HesB-like domain |
| 13 | c2qgoA_
|
|
 |
97.6 |
13 |
PDB header:biosynthetic protein
Chain: A: PDB Molecule:putative fe-s biosynthesis protein;
PDBTitle: crystal structure of a putative fe-s biosynthesis protein from2 lactobacillus acidophilus
|
| 14 | c3lnoA_
|
|
 |
96.1 |
16 |
PDB header:unknown function
Chain: A: PDB Molecule:putative uncharacterized protein;
PDBTitle: crystal structure of domain of unknown function duf59 from2 bacillus anthracis
|
| 15 | d2cu6a1
|
|
 |
93.9 |
19 |
Fold:Alpha-lytic protease prodomain-like
Superfamily:Fe-S cluster assembly (FSCA) domain-like
Family:PaaD-like |
| 16 | d1uwda_
|
|
 |
91.4 |
14 |
Fold:Alpha-lytic protease prodomain-like
Superfamily:Fe-S cluster assembly (FSCA) domain-like
Family:PaaD-like |
| 17 | c3qauA_
|
|
 |
63.9 |
13 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:3-hydroxy-3-methylglutaryl-coenzyme a reductase;
PDBTitle: 3-hydroxy-3-methylglutaryl-coenzyme a reductase from streptococcus2 pneumoniae
|
| 18 | c1r7iB_
|
|
 |
59.5 |
7 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:3-hydroxy-3-methylglutaryl-coenzyme a reductase;
PDBTitle: hmg-coa reductase from p. mevalonii, native structure at 2.2 angstroms2 resolution.
|
| 19 | d2c42a2
|
|
 |
56.1 |
25 |
Fold:Thiamin diphosphate-binding fold (THDP-binding)
Superfamily:Thiamin diphosphate-binding fold (THDP-binding)
Family:PFOR PP module |
| 20 | d1xg8a_
|
|
 |
50.6 |
12 |
Fold:Thioredoxin fold
Superfamily:Thioredoxin-like
Family:YuzD-like |
| 21 | d1t82a_ |
|
not modelled |
43.6 |
22 |
Fold:Thioesterase/thiol ester dehydrase-isomerase
Superfamily:Thioesterase/thiol ester dehydrase-isomerase
Family:PaaI/YdiI-like |
| 22 | c3lwaA_ |
|
not modelled |
40.2 |
20 |
PDB header:isomerase
Chain: A: PDB Molecule:secreted thiol-disulfide isomerase;
PDBTitle: the crystal structure of a secreted thiol-disulfide2 isomerase from corynebacterium glutamicum to 1.75a
|
| 23 | c2c3yA_ |
|
not modelled |
33.0 |
24 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:pyruvate-ferredoxin oxidoreductase;
PDBTitle: crystal structure of the radical form of2 pyruvate:ferredoxin oxidoreductase from desulfovibrio3 africanus
|
| 24 | d2a0sa1 |
|
not modelled |
32.5 |
16 |
Fold:T-fold
Superfamily:Tetrahydrobiopterin biosynthesis enzymes-like
Family:6-pyruvoyl tetrahydropterin synthase |
| 25 | d1xn7a_ |
|
not modelled |
31.6 |
33 |
Fold:DNA/RNA-binding 3-helical bundle
Superfamily:"Winged helix" DNA-binding domain
Family:Hypothetical protein YhgG |
| 26 | d1wp0a1 |
|
not modelled |
29.4 |
8 |
Fold:Thioredoxin fold
Superfamily:Thioredoxin-like
Family:Glutathione peroxidase-like |
| 27 | d1qxha_ |
|
not modelled |
29.2 |
10 |
Fold:Thioredoxin fold
Superfamily:Thioredoxin-like
Family:Glutathione peroxidase-like |
| 28 | c2rmlA_ |
|
not modelled |
28.3 |
28 |
PDB header:hydrolase
Chain: A: PDB Molecule:copper-transporting p-type atpase copa;
PDBTitle: solution structure of the n-terminal soluble domains of2 bacillus subtilis copa
|
| 29 | c1yjrA_ |
|
not modelled |
26.0 |
32 |
PDB header:hydrolase
Chain: A: PDB Molecule:copper-transporting atpase 1;
PDBTitle: solution structure of the apo form of the sixth soluble2 domain a69p mutant of menkes protein
|
| 30 | d1e2ya_ |
|
not modelled |
22.9 |
7 |
Fold:Thioredoxin fold
Superfamily:Thioredoxin-like
Family:Glutathione peroxidase-like |
| 31 | d1y13a_ |
|
not modelled |
21.7 |
15 |
Fold:T-fold
Superfamily:Tetrahydrobiopterin biosynthesis enzymes-like
Family:6-pyruvoyl tetrahydropterin synthase |
| 32 | d1j2ga2 |
|
not modelled |
20.4 |
24 |
Fold:T-fold
Superfamily:Tetrahydrobiopterin biosynthesis enzymes-like
Family:Urate oxidase (uricase) |
| 33 | c1bcrA_ |
|
not modelled |
20.1 |
13 |
PDB header:hydrolase/hydrolase inhibitor
Chain: A: PDB Molecule:serine carboxypeptidase ii;
PDBTitle: complex of the wheat serine carboxypeptidase, cpdw-ii, with the2 microbial peptide aldehyde inhibitor, antipain, and arginine at room3 temperature
|
| 34 | c2dhmA_ |
|
not modelled |
19.7 |
20 |
PDB header:protein binding
Chain: A: PDB Molecule:protein bola;
PDBTitle: solution structure of the bola protein from escherichia coli
|
| 35 | d1s6ua_ |
|
not modelled |
19.7 |
18 |
Fold:Ferredoxin-like
Superfamily:HMA, heavy metal-associated domain
Family:HMA, heavy metal-associated domain |
| 36 | c2rogA_ |
|
not modelled |
19.1 |
35 |
PDB header:metal binding protein
Chain: A: PDB Molecule:heavy metal binding protein;
PDBTitle: solution structure of thermus thermophilus hb8 ttha17182 protein in living e. coli cells
|
| 37 | c3e8pA_ |
|
not modelled |
18.9 |
16 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:uncharacterized protein;
PDBTitle: crystal structure of the protein q8e9m7 from shewanella2 oneidensis related to thioesterase superfamily. northeast3 structural genomics consortium target sor246.
|
| 38 | d2ibaa2 |
|
not modelled |
18.6 |
20 |
Fold:T-fold
Superfamily:Tetrahydrobiopterin biosynthesis enzymes-like
Family:Urate oxidase (uricase) |
| 39 | c3ixrA_ |
|
not modelled |
18.2 |
9 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:bacterioferritin comigratory protein;
PDBTitle: crystal structure of xylella fastidiosa prxq c47s mutant
|
| 40 | c2rliA_ |
|
not modelled |
17.7 |
8 |
PDB header:metal transport
Chain: A: PDB Molecule:sco2 protein homolog, mitochondrial;
PDBTitle: solution structure of cu(i) human sco2
|
| 41 | c2k5eA_ |
|
not modelled |
16.8 |
29 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:uncharacterized protein;
PDBTitle: solution structure of putative uncharacterized protein2 gsu1278 from methanocaldococcus jannaschii, northeast3 structural genomics consortium (nesg) target gsr195
|
| 42 | c1y8oA_ |
|
not modelled |
16.5 |
15 |
PDB header:transferase
Chain: A: PDB Molecule:[pyruvate dehydrogenase [lipoamide]] kinase isozyme 3;
PDBTitle: crystal structure of the pdk3-l2 complex
|
| 43 | d2yzca2 |
|
not modelled |
15.7 |
12 |
Fold:T-fold
Superfamily:Tetrahydrobiopterin biosynthesis enzymes-like
Family:Urate oxidase (uricase) |
| 44 | c1gjvA_ |
|
not modelled |
15.7 |
19 |
PDB header:transferase
Chain: A: PDB Molecule:[3-methyl-2-oxobutanoate dehydrogenase
PDBTitle: branched-chain alpha-ketoacid dehydrogenase kinase (bck)2 complxed with atp-gamma-s
|
| 45 | c2npbA_ |
|
not modelled |
15.4 |
14 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:selenoprotein w;
PDBTitle: nmr solution structure of mouse selw
|
| 46 | c1gxsC_ |
|
not modelled |
14.8 |
16 |
PDB header:lyase
Chain: C: PDB Molecule:p-(s)-hydroxymandelonitrile lyase chain a;
PDBTitle: crystal structure of hydroxynitrile lyase from sorghum2 bicolor in complex with inhibitor benzoic acid: a novel3 cyanogenic enzyme
|
| 47 | c2kt2A_ |
|
not modelled |
14.6 |
21 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:mercuric reductase;
PDBTitle: structure of nmera, the n-terminal hma domain of tn501 mercuric2 reductase
|
| 48 | c2q8fA_ |
|
not modelled |
14.0 |
15 |
PDB header:transferase
Chain: A: PDB Molecule:[pyruvate dehydrogenase [lipoamide]] kinase isozyme 1;
PDBTitle: structure of pyruvate dehydrogenase kinase isoform 1
|
| 49 | d1r9fa_ |
|
not modelled |
13.8 |
21 |
Fold:Tombusvirus P19 core protein, VP19
Superfamily:Tombusvirus P19 core protein, VP19
Family:Tombusvirus P19 core protein, VP19 |
| 50 | d1q8la_ |
|
not modelled |
13.7 |
19 |
Fold:Ferredoxin-like
Superfamily:HMA, heavy metal-associated domain
Family:HMA, heavy metal-associated domain |
| 51 | c2b7kD_ |
|
not modelled |
13.6 |
11 |
PDB header:metal binding protein
Chain: D: PDB Molecule:sco1 protein;
PDBTitle: crystal structure of yeast sco1
|
| 52 | d2b7ka1 |
|
not modelled |
13.4 |
8 |
Fold:Thioredoxin fold
Superfamily:Thioredoxin-like
Family:Glutathione peroxidase-like |
| 53 | c2k53A_ |
|
not modelled |
13.2 |
38 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:a3dk08 protein;
PDBTitle: nmr solution structure of a3dk08 protein from clostridium2 thermocellum: northeast structural genomics consortium3 target cmr9
|
| 54 | d1rpua_ |
|
not modelled |
13.0 |
21 |
Fold:Tombusvirus P19 core protein, VP19
Superfamily:Tombusvirus P19 core protein, VP19
Family:Tombusvirus P19 core protein, VP19 |
| 55 | c1rpuA_ |
|
not modelled |
13.0 |
21 |
PDB header:rna binding protein/rna
Chain: A: PDB Molecule:19 kda protein;
PDBTitle: crystal structure of cirv p19 bound to sirna
|
| 56 | c3lgbB_ |
|
not modelled |
12.7 |
14 |
PDB header:transferase
Chain: B: PDB Molecule:dna primase large subunit;
PDBTitle: crystal structure of the fe-s domain of the yeast dna primase
|
| 57 | c3cynC_ |
|
not modelled |
12.6 |
20 |
PDB header:oxidoreductase
Chain: C: PDB Molecule:probable glutathione peroxidase 8;
PDBTitle: the structure of human gpx8
|
| 58 | c1j2gC_ |
|
not modelled |
12.6 |
24 |
PDB header:oxidoreductase
Chain: C: PDB Molecule:uricase;
PDBTitle: crystal structure of urate oxidase from bacillus sp. tb-90 co-2 crystallized with 8-azaxanthine
|
| 59 | c2jszA_ |
|
not modelled |
12.6 |
7 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:probable thiol peroxidase;
PDBTitle: solution structure of tpx in the reduced state
|
| 60 | c2v1mA_ |
|
not modelled |
12.5 |
15 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:glutathione peroxidase;
PDBTitle: crystal structure of schistosoma mansoni glutathione2 peroxidase
|
| 61 | d1owxa_ |
|
not modelled |
12.2 |
17 |
Fold:Ferredoxin-like
Superfamily:RNA-binding domain, RBD
Family:Canonical RBD |
| 62 | c3gknA_ |
|
not modelled |
12.1 |
7 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:bacterioferritin comigratory protein;
PDBTitle: insights into the alkyl peroxide reduction activity of xanthomonas2 campestris bacterioferritin comigratory protein from the trapped3 intermediate/ligand complex structures
|
| 63 | c1zyeL_ |
|
not modelled |
12.0 |
9 |
PDB header:oxidoreductase
Chain: L: PDB Molecule:thioredoxin-dependent peroxide reductase;
PDBTitle: crystal strucutre analysis of bovine mitochondrial peroxiredoxin iii
|
| 64 | c3qq5A_ |
|
not modelled |
12.0 |
17 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:small gtp-binding protein;
PDBTitle: crystal structure of the [fefe]-hydrogenase maturation protein hydf
|
| 65 | c1r56H_ |
|
not modelled |
11.9 |
20 |
PDB header:oxidoreductase
Chain: H: PDB Molecule:uricase;
PDBTitle: uncomplexed urate oxidase from aspergillus flavus
|
| 66 | c2yzbA_ |
|
not modelled |
11.5 |
12 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:uricase;
PDBTitle: crystal structure of uricase from arthrobacter globiformis2 in complex with uric acid (substrate)
|
| 67 | d1q98a_ |
|
not modelled |
11.5 |
6 |
Fold:Thioredoxin fold
Superfamily:Thioredoxin-like
Family:Glutathione peroxidase-like |
| 68 | d1afia_ |
|
not modelled |
11.1 |
23 |
Fold:Ferredoxin-like
Superfamily:HMA, heavy metal-associated domain
Family:HMA, heavy metal-associated domain |
| 69 | d1o73a_ |
|
not modelled |
10.5 |
31 |
Fold:Thioredoxin fold
Superfamily:Thioredoxin-like
Family:Glutathione peroxidase-like |
| 70 | d2aw0a_ |
|
not modelled |
10.4 |
35 |
Fold:Ferredoxin-like
Superfamily:HMA, heavy metal-associated domain
Family:HMA, heavy metal-associated domain |
| 71 | c2gvsA_ |
|
not modelled |
10.2 |
21 |
PDB header:lipid binding protein
Chain: A: PDB Molecule:chemosensory protein csp-sg4;
PDBTitle: nmr solution structure of cspsg4
|
| 72 | d1kx9b_ |
|
not modelled |
9.9 |
17 |
Fold:alpha-alpha superhelix
Superfamily:Chemosensory protein Csp2
Family:Chemosensory protein Csp2 |
| 73 | c3me5A_ |
|
not modelled |
9.7 |
22 |
PDB header:transferase
Chain: A: PDB Molecule:cytosine-specific methyltransferase;
PDBTitle: crystal structure of putative dna cytosine methylase from shigella2 flexneri 2a str. 301
|
| 74 | c2kyzA_ |
|
not modelled |
9.7 |
30 |
PDB header:metal binding protein
Chain: A: PDB Molecule:heavy metal binding protein;
PDBTitle: nmr structure of heavy metal binding protein tm0320 from thermotoga2 maritima
|
| 75 | c2ga7A_ |
|
not modelled |
9.6 |
31 |
PDB header:hydrolase
Chain: A: PDB Molecule:copper-transporting atpase 1;
PDBTitle: solution structure of the copper(i) form of the third metal-2 binding domain of atp7a protein (menkes disease protein)
|
| 76 | d2cx4a1 |
|
not modelled |
9.4 |
13 |
Fold:Thioredoxin fold
Superfamily:Thioredoxin-like
Family:Glutathione peroxidase-like |
| 77 | d2ggpb1 |
|
not modelled |
9.3 |
29 |
Fold:Ferredoxin-like
Superfamily:HMA, heavy metal-associated domain
Family:HMA, heavy metal-associated domain |
| 78 | d1n8va_ |
|
not modelled |
9.2 |
17 |
Fold:alpha-alpha superhelix
Superfamily:Chemosensory protein Csp2
Family:Chemosensory protein Csp2 |
| 79 | d1osda_ |
|
not modelled |
9.1 |
19 |
Fold:Ferredoxin-like
Superfamily:HMA, heavy metal-associated domain
Family:HMA, heavy metal-associated domain |
| 80 | d2a4va1 |
|
not modelled |
9.1 |
10 |
Fold:Thioredoxin fold
Superfamily:Thioredoxin-like
Family:Glutathione peroxidase-like |
| 81 | c3dwvB_ |
|
not modelled |
9.0 |
12 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:glutathione peroxidase-like protein;
PDBTitle: glutathione peroxidase-type tryparedoxin peroxidase,2 oxidized form
|
| 82 | d1dosa_ |
|
not modelled |
8.8 |
17 |
Fold:TIM beta/alpha-barrel
Superfamily:Aldolase
Family:Class II FBP aldolase |
| 83 | d1xzoa1 |
|
not modelled |
8.6 |
16 |
Fold:Thioredoxin fold
Superfamily:Thioredoxin-like
Family:Glutathione peroxidase-like |
| 84 | d1gp1a_ |
|
not modelled |
8.5 |
8 |
Fold:Thioredoxin fold
Superfamily:Thioredoxin-like
Family:Glutathione peroxidase-like |
| 85 | d1p6ta1 |
|
not modelled |
8.3 |
29 |
Fold:Ferredoxin-like
Superfamily:HMA, heavy metal-associated domain
Family:HMA, heavy metal-associated domain |
| 86 | d1rutx3 |
|
not modelled |
8.2 |
43 |
Fold:Glucocorticoid receptor-like (DNA-binding domain)
Superfamily:Glucocorticoid receptor-like (DNA-binding domain)
Family:LIM domain |
| 87 | c2l9wA_ |
|
not modelled |
7.8 |
12 |
PDB header:splicing, rna binding protein
Chain: A: PDB Molecule:u4/u6 snrna-associated-splicing factor prp24;
PDBTitle: solution structure of the c-terminal domain of prp24
|
| 88 | c2ywiA_ |
|
not modelled |
7.7 |
13 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:hypothetical conserved protein;
PDBTitle: crystal structure of uncharacterized conserved protein from2 geobacillus kaustophilus
|
| 89 | c2he3A_ |
|
not modelled |
7.7 |
15 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:glutathione peroxidase 2;
PDBTitle: crystal structure of the selenocysteine to cysteine mutant of human2 glutathionine peroxidase 2 (gpx2)
|
| 90 | d1u9ya1 |
|
not modelled |
7.6 |
16 |
Fold:PRTase-like
Superfamily:PRTase-like
Family:Phosphoribosylpyrophosphate synthetase-like |
| 91 | d1yexa1 |
|
not modelled |
7.6 |
8 |
Fold:Thioredoxin fold
Superfamily:Thioredoxin-like
Family:Glutathione peroxidase-like |
| 92 | c3kebB_ |
|
not modelled |
7.4 |
9 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:probable thiol peroxidase;
PDBTitle: thiol peroxidase from chromobacterium violaceum
|
| 93 | c3l9qB_ |
|
not modelled |
7.3 |
19 |
PDB header:transferase
Chain: B: PDB Molecule:dna primase large subunit;
PDBTitle: crystal structure of human polymerase alpha-primase p58 iron-sulfur2 cluster domain
|
| 94 | c2gcfA_ |
|
not modelled |
7.1 |
26 |
PDB header:hydrolase
Chain: A: PDB Molecule:cation-transporting atpase pacs;
PDBTitle: solution structure of the n-terminal domain of the coppper(i) atpase2 pacs in its apo form
|
| 95 | c3i24B_ |
|
not modelled |
7.1 |
20 |
PDB header:hydrolase
Chain: B: PDB Molecule:hit family hydrolase;
PDBTitle: crystal structure of a hit family hydrolase protein from2 vibrio fischeri. northeast structural genomics consortium3 target id vfr176
|
| 96 | c3dxsX_ |
|
not modelled |
7.0 |
19 |
PDB header:hydrolase
Chain: X: PDB Molecule:copper-transporting atpase ran1;
PDBTitle: crystal structure of a copper binding domain from hma7, a p-2 type atpase
|
| 97 | c3ha9A_ |
|
not modelled |
7.0 |
22 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:uncharacterized thioredoxin-like protein;
PDBTitle: the 1.7a crystal structure of a thioredoxin-like protein from2 aeropyrum pernix
|
| 98 | d1xvwa1 |
|
not modelled |
7.0 |
10 |
Fold:Thioredoxin fold
Superfamily:Thioredoxin-like
Family:Glutathione peroxidase-like |
| 99 | c3eurA_ |
|
not modelled |
6.9 |
16 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:uncharacterized protein;
PDBTitle: crystal structure of the c-terminal domain of uncharacterized protein2 from bacteroides fragilis nctc 9343
|