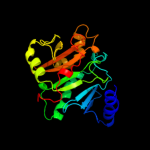
| 1 | d2pofa1 |
|
 |
100.0 |
96 |
Fold:HIT-like
Superfamily:HIT-like
Family:CDH-like |
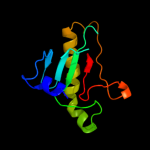
| 2 | c3n1tE_ |
|
 |
96.2 |
12 |
PDB header:hydrolase
Chain: E: PDB Molecule:hit-like protein hint;
PDBTitle: crystal structure of the h101a mutant echint gmp complex
|
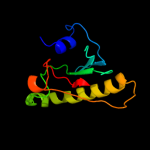
| 3 | c1xquA_ |
|
 |
95.9 |
16 |
PDB header:hydrolase
Chain: A: PDB Molecule:hit family hydrolase;
PDBTitle: hit family hydrolase from clostridium thermocellum cth-393
|
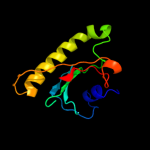
| 4 | d1xqua_ |
|
 |
95.9 |
16 |
Fold:HIT-like
Superfamily:HIT-like
Family:HIT (HINT, histidine triad) family of protein kinase-interacting proteins |
| 5 | c1emsB_ |
|
 |
95.9 |
17 |
PDB header:antitumor protein
Chain: B: PDB Molecule:nit-fragile histidine triad fusion protein;
PDBTitle: crystal structure of the c. elegans nitfhit protein
|
| 6 | d1emsa1 |
|
 |
95.8 |
15 |
Fold:HIT-like
Superfamily:HIT-like
Family:HIT (HINT, histidine triad) family of protein kinase-interacting proteins |
| 7 | c3anoA_ |
|
 |
95.7 |
14 |
PDB header:transferase
Chain: A: PDB Molecule:ap-4-a phosphorylase;
PDBTitle: crystal structure of a novel diadenosine 5',5'''-p1,p4-tetraphosphate2 phosphorylase from mycobacterium tuberculosis h37rv
|
| 8 | d1rzya_ |
|
 |
95.5 |
12 |
Fold:HIT-like
Superfamily:HIT-like
Family:HIT (HINT, histidine triad) family of protein kinase-interacting proteins |
| 9 | c3lb5B_ |
|
 |
95.4 |
13 |
PDB header:cell cycle
Chain: B: PDB Molecule:hit-like protein involved in cell-cycle regulation;
PDBTitle: crystal structure of hit-like protein involved in cell-cycle2 regulation from bartonella henselae with unknown ligand
|
| 10 | d1kpfa_ |
|
 |
95.4 |
14 |
Fold:HIT-like
Superfamily:HIT-like
Family:HIT (HINT, histidine triad) family of protein kinase-interacting proteins |
| 11 | c3oj7A_ |
|
 |
95.1 |
24 |
PDB header:metal binding protein
Chain: A: PDB Molecule:putative histidine triad family protein;
PDBTitle: crystal structure of a histidine triad family protein from entamoeba2 histolytica, bound to sulfate
|
| 12 | d1fita_ |
|
 |
94.9 |
15 |
Fold:HIT-like
Superfamily:HIT-like
Family:HIT (HINT, histidine triad) family of protein kinase-interacting proteins |
| 13 | c3ksvA_ |
|
 |
94.3 |
17 |
PDB header:unknown function
Chain: A: PDB Molecule:uncharacterized protein;
PDBTitle: hypothetical protein from leishmania major
|
| 14 | d1vlra1 |
|
 |
93.8 |
22 |
Fold:HIT-like
Superfamily:HIT-like
Family:mRNA decapping enzyme DcpS C-terminal domain |
| 15 | c2eo4A_ |
|
 |
93.7 |
17 |
PDB header:hydrolase
Chain: A: PDB Molecule:150aa long hypothetical histidine triad nucleotide-binding
PDBTitle: crystal structure of hypothetical histidine triad nucleotide-binding2 protein st2152 from sulfolobus tokodaii strain7
|
| 16 | c3l7xA_ |
|
 |
93.7 |
16 |
PDB header:cell cycle
Chain: A: PDB Molecule:putative hit-like protein involved in cell-cycle
PDBTitle: the crystal structure of smu.412c from streptococcus mutans ua159
|
| 17 | c3r6fA_ |
|
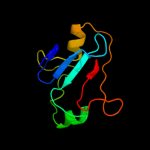 |
93.5 |
13 |
PDB header:hydrolase
Chain: A: PDB Molecule:hit family protein;
PDBTitle: crystal structure of a zinc-containing hit family protein from2 encephalitozoon cuniculi
|
| 18 | d1z84a2 |
|
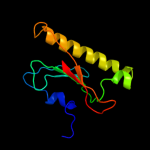 |
93.5 |
11 |
Fold:HIT-like
Superfamily:HIT-like
Family:Hexose-1-phosphate uridylyltransferase |
| 19 | d3bl9a1 |
|
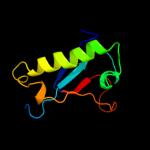 |
93.1 |
19 |
Fold:HIT-like
Superfamily:HIT-like
Family:mRNA decapping enzyme DcpS C-terminal domain |
| 20 | c3bl9B_ |
|
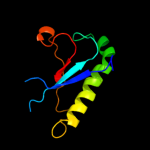 |
92.7 |
22 |
PDB header:hydrolase
Chain: B: PDB Molecule:scavenger mrna-decapping enzyme dcps;
PDBTitle: synthetic gene encoded dcps bound to inhibitor dg157493
|
| 21 | c3o0mB_ |
|
not modelled |
92.2 |
14 |
PDB header:hydrolase
Chain: B: PDB Molecule:hit family protein;
PDBTitle: crystal structure of a zn-bound histidine triad family protein from2 mycobacterium smegmatis
|
| 22 | c1xmlA_ |
|
not modelled |
91.5 |
20 |
PDB header:chaperone
Chain: A: PDB Molecule:heat shock-like protein 1;
PDBTitle: structure of human dcps
|
| 23 | c3imiB_ |
|
not modelled |
90.4 |
19 |
PDB header:structural genomics, unknown function
Chain: B: PDB Molecule:hit family protein;
PDBTitle: 2.01 angstrom resolution crystal structure of a hit family protein2 from bacillus anthracis str. 'ames ancestor'
|
| 24 | d1y23a_ |
|
not modelled |
89.0 |
17 |
Fold:HIT-like
Superfamily:HIT-like
Family:HIT (HINT, histidine triad) family of protein kinase-interacting proteins |
| 25 | d1guqa2 |
|
not modelled |
88.4 |
15 |
Fold:HIT-like
Superfamily:HIT-like
Family:Hexose-1-phosphate uridylyltransferase |
| 26 | c3p0tB_ |
|
not modelled |
88.1 |
17 |
PDB header:unknown function
Chain: B: PDB Molecule:uncharacterized protein;
PDBTitle: crystal structure of an hit-like protein from mycobacterium2 paratuberculosis
|
| 27 | c3i24B_ |
|
not modelled |
85.1 |
11 |
PDB header:hydrolase
Chain: B: PDB Molecule:hit family hydrolase;
PDBTitle: crystal structure of a hit family hydrolase protein from2 vibrio fischeri. northeast structural genomics consortium3 target id vfr176
|
| 28 | d2oika1 |
|
not modelled |
84.6 |
17 |
Fold:HIT-like
Superfamily:HIT-like
Family:HIT (HINT, histidine triad) family of protein kinase-interacting proteins |
| 29 | c1zwjA_ |
|
not modelled |
80.5 |
11 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:putative galactose-1-phosphate uridyl transferase;
PDBTitle: x-ray structure of galt-like protein from arabidopsis thaliana2 at5g18200
|
| 30 | c3nrdB_ |
|
not modelled |
74.4 |
13 |
PDB header:nucleotide binding protein
Chain: B: PDB Molecule:histidine triad (hit) protein;
PDBTitle: crystal structure of a histidine triad (hit) protein (smc02904) from2 sinorhizobium meliloti 1021 at 2.06 a resolution
|
| 31 | c3i4sB_ |
|
not modelled |
71.9 |
19 |
PDB header:hydrolase
Chain: B: PDB Molecule:histidine triad protein;
PDBTitle: crystal structure of histidine triad protein blr8122 from2 bradyrhizobium japonicum
|
| 32 | c3oheA_ |
|
not modelled |
68.4 |
13 |
PDB header:hydrolase
Chain: A: PDB Molecule:histidine triad (hit) protein;
PDBTitle: crystal structure of a histidine triad protein (maqu_1709) from2 marinobacter aquaeolei vt8 at 1.20 a resolution
|
| 33 | c1gupC_ |
|
not modelled |
61.7 |
15 |
PDB header:nucleotidyltransferase
Chain: C: PDB Molecule:galactose-1-phosphate uridylyltransferase;
PDBTitle: structure of nucleotidyltransferase complexed with udp-2 galactose
|
| 34 | d1z84a1 |
|
not modelled |
43.8 |
11 |
Fold:HIT-like
Superfamily:HIT-like
Family:Hexose-1-phosphate uridylyltransferase |
| 35 | d2soba_ |
|
not modelled |
36.0 |
19 |
Fold:OB-fold
Superfamily:Staphylococcal nuclease
Family:Staphylococcal nuclease |
| 36 | c3bdlA_ |
|
not modelled |
35.7 |
26 |
PDB header:hydrolase
Chain: A: PDB Molecule:staphylococcal nuclease domain-containing
PDBTitle: crystal structure of a truncated human tudor-sn
|
| 37 | d1rkna_ |
|
not modelled |
30.0 |
22 |
Fold:OB-fold
Superfamily:Staphylococcal nuclease
Family:Staphylococcal nuclease |
| 38 | c2jvaA_ |
|
not modelled |
22.5 |
9 |
PDB header:hydrolase
Chain: A: PDB Molecule:peptidyl-trna hydrolase domain protein;
PDBTitle: nmr solution structure of peptidyl-trna hydrolase domain protein from2 pseudomonas syringae pv. tomato. northeast structural genomics3 consortium target psr211
|
| 39 | c3ngbI_ |
|
not modelled |
21.8 |
44 |
PDB header:viral protein/immune system
Chain: I: PDB Molecule:envelope glycoprotein gp160;
PDBTitle: crystal structure of broadly and potently neutralizing antibody vrc012 in complex with hiv-1 gp120
|
| 40 | d1vdla_ |
|
not modelled |
20.6 |
18 |
Fold:RuvA C-terminal domain-like
Superfamily:UBA-like
Family:UBA domain |
| 41 | d1qpma_ |
|
not modelled |
19.2 |
13 |
Fold:Putative DNA-binding domain
Superfamily:Putative DNA-binding domain
Family:Excisionase-like |
| 42 | d1tnsa_ |
|
not modelled |
19.0 |
9 |
Fold:Putative DNA-binding domain
Superfamily:Putative DNA-binding domain
Family:Excisionase-like |
| 43 | d2amxa1 |
|
not modelled |
17.8 |
10 |
Fold:TIM beta/alpha-barrel
Superfamily:Metallo-dependent hydrolases
Family:Adenosine/AMP deaminase |
| 44 | c3gwhB_ |
|
not modelled |
15.1 |
18 |
PDB header:transcription
Chain: B: PDB Molecule:transcriptional antiterminator (bglg family);
PDBTitle: crystallographic ab initio protein solution far below atomic2 resolution
|
| 45 | c3dnlB_ |
|
not modelled |
14.8 |
29 |
PDB header:viral protein
Chain: B: PDB Molecule:hiv-1 envelope glycoprotein gp120;
PDBTitle: molecular structure for the hiv-1 gp120 trimer in the b12-2 bound state
|
| 46 | c1ql1A_ |
|
not modelled |
12.9 |
32 |
PDB header:virus
Chain: A: PDB Molecule:pf1 bacteriophage coat protein b;
PDBTitle: inovirus (filamentous bacteriophage) strain pf1 major coat2 protein assembly
|
| 47 | d1tafa_ |
|
not modelled |
12.8 |
15 |
Fold:Histone-fold
Superfamily:Histone-fold
Family:TBP-associated factors, TAFs |
| 48 | c3jwdA_ |
|
not modelled |
12.3 |
40 |
PDB header:viral protein
Chain: A: PDB Molecule:hiv-1 gp120 envelope glycoprotein;
PDBTitle: structure of hiv-1 gp120 with gp41-interactive region: layered2 architecture and basis of conformational mobility
|
| 49 | d1jz8a1 |
|
not modelled |
12.0 |
38 |
Fold:Immunoglobulin-like beta-sandwich
Superfamily:beta-Galactosidase/glucuronidase domain
Family:beta-Galactosidase/glucuronidase domain |
| 50 | d1snoa_ |
|
not modelled |
11.7 |
22 |
Fold:OB-fold
Superfamily:Staphylococcal nuclease
Family:Staphylococcal nuclease |
| 51 | c2i45C_ |
|
not modelled |
11.5 |
22 |
PDB header:structural genomics, unknown function
Chain: C: PDB Molecule:hypothetical protein;
PDBTitle: crystal structure of protein nmb1881 from neisseria meningitidis
|
| 52 | d1yq2a2 |
|
not modelled |
11.1 |
15 |
Fold:Immunoglobulin-like beta-sandwich
Superfamily:beta-Galactosidase/glucuronidase domain
Family:beta-Galactosidase/glucuronidase domain |
| 53 | d1mr0a_ |
|
not modelled |
11.1 |
21 |
Fold:Knottins (small inhibitors, toxins, lectins)
Superfamily:Agouti-related protein
Family:Agouti-related protein |
| 54 | d1k8kd2 |
|
not modelled |
10.2 |
18 |
Fold:Secretion chaperone-like
Superfamily:Arp2/3 complex subunits
Family:Arp2/3 complex subunits |
| 55 | c2i34B_ |
|
not modelled |
9.7 |
14 |
PDB header:hydrolase
Chain: B: PDB Molecule:acid phosphatase;
PDBTitle: the crystal structure of class c acid phosphatase from bacillus2 anthracis with tungstate bound
|
| 56 | c3dwlI_ |
|
not modelled |
9.6 |
18 |
PDB header:structural protein
Chain: I: PDB Molecule:actin-related protein 2/3 complex subunit 2;
PDBTitle: crystal structure of fission yeast arp2/3 complex lacking the arp22 subunit
|
| 57 | d2fnaa1 |
|
not modelled |
9.5 |
9 |
Fold:DNA/RNA-binding 3-helical bundle
Superfamily:"Winged helix" DNA-binding domain
Family:Helicase DNA-binding domain |
| 58 | c2k21A_ |
|
not modelled |
9.3 |
20 |
PDB header:membrane protein
Chain: A: PDB Molecule:potassium voltage-gated channel subfamily e
PDBTitle: nmr structure of human kcne1 in lmpg micelles at ph 6.0 and2 40 degree c
|
| 59 | d1es6a2 |
|
not modelled |
9.0 |
43 |
Fold:EV matrix protein
Superfamily:EV matrix protein
Family:EV matrix protein |
| 60 | c2rrlA_ |
|
not modelled |
8.8 |
20 |
PDB header:protein transport
Chain: A: PDB Molecule:flagellar hook-length control protein;
PDBTitle: solution structure of the c-terminal domain of the flik
|
| 61 | c3u1dA_ |
|
not modelled |
8.3 |
24 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:uncharacterized protein;
PDBTitle: the structure of a protein with a gntr superfamily winged-helix-turn-2 helix domain from halomicrobium mukohataei.
|
| 62 | c2k2wA_ |
|
not modelled |
8.2 |
25 |
PDB header:cell cycle
Chain: A: PDB Molecule:recombination and dna repair protein;
PDBTitle: second brct domain of nbs1
|
| 63 | c3hx6A_ |
|
not modelled |
7.4 |
28 |
PDB header:cell adhesion
Chain: A: PDB Molecule:type 4 fimbrial biogenesis protein pily1;
PDBTitle: crystal structure of pseudomonas aeruginosa pily1 c-terminal2 domain
|
| 64 | d2glia1 |
|
not modelled |
7.2 |
30 |
Fold:beta-beta-alpha zinc fingers
Superfamily:beta-beta-alpha zinc fingers
Family:Classic zinc finger, C2H2 |
| 65 | d2b1xa1 |
|
not modelled |
7.0 |
15 |
Fold:ISP domain
Superfamily:ISP domain
Family:Ring hydroxylating alpha subunit ISP domain |
| 66 | d1a4ma_ |
|
not modelled |
6.9 |
19 |
Fold:TIM beta/alpha-barrel
Superfamily:Metallo-dependent hydrolases
Family:Adenosine/AMP deaminase |
| 67 | d1j26a_ |
|
not modelled |
6.6 |
16 |
Fold:dsRBD-like
Superfamily:Peptidyl-tRNA hydrolase domain-like
Family:Peptidyl-tRNA hydrolase domain |
| 68 | d2nxya1 |
|
not modelled |
6.3 |
29 |
Fold:gp120 core
Superfamily:gp120 core
Family:gp120 core |
| 69 | c2p9lD_ |
|
not modelled |
6.3 |
18 |
PDB header:structural protein
Chain: D: PDB Molecule:actin-related protein 2/3 complex subunit 2;
PDBTitle: crystal structure of bovine arp2/3 complex
|
| 70 | c2iirJ_ |
|
not modelled |
6.2 |
26 |
PDB header:transferase
Chain: J: PDB Molecule:acetate kinase;
PDBTitle: acetate kinase from a hypothermophile thermotoga maritima
|
| 71 | c3rysA_ |
|
not modelled |
6.2 |
24 |
PDB header:hydrolase
Chain: A: PDB Molecule:adenosine deaminase 1;
PDBTitle: the crystal structure of adenine deaminase (aaur1117) from2 arthrobacter aurescens
|
| 72 | d1hqia_ |
|
not modelled |
6.2 |
11 |
Fold:Monooxygenase (hydroxylase) regulatory protein
Superfamily:Monooxygenase (hydroxylase) regulatory protein
Family:Monooxygenase (hydroxylase) regulatory protein |
| 73 | d2b7ea1 |
|
not modelled |
6.1 |
45 |
Fold:Another 3-helical bundle
Superfamily:FF domain
Family:FF domain |
| 74 | d2ihoa2 |
|
not modelled |
6.1 |
14 |
Fold:Cysteine proteinases
Superfamily:Cysteine proteinases
Family:MOA C-terminal domain-like |
| 75 | d1t0ga_ |
|
not modelled |
6.0 |
19 |
Fold:Cytochrome b5-like heme/steroid binding domain
Superfamily:Cytochrome b5-like heme/steroid binding domain
Family:Steroid-binding domain |
| 76 | d1vfla1 |
|
not modelled |
6.0 |
19 |
Fold:TIM beta/alpha-barrel
Superfamily:Metallo-dependent hydrolases
Family:Adenosine/AMP deaminase |
| 77 | d1iufa1 |
|
not modelled |
6.0 |
15 |
Fold:DNA/RNA-binding 3-helical bundle
Superfamily:Homeodomain-like
Family:Centromere-binding |
| 78 | d1d2za_ |
|
not modelled |
5.9 |
30 |
Fold:DEATH domain
Superfamily:DEATH domain
Family:DEATH domain, DD |
| 79 | c2l1nA_ |
|
not modelled |
5.9 |
27 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:uncharacterized protein;
PDBTitle: solution nmr structure of the protein yp_399305.1
|
| 80 | d2bf2a1 |
|
not modelled |
5.6 |
19 |
Fold:Monooxygenase (hydroxylase) regulatory protein
Superfamily:Monooxygenase (hydroxylase) regulatory protein
Family:Monooxygenase (hydroxylase) regulatory protein |
| 81 | d1k8ke_ |
|
not modelled |
5.6 |
27 |
Fold:Arp2/3 complex 21 kDa subunit ARPC3
Superfamily:Arp2/3 complex 21 kDa subunit ARPC3
Family:Arp2/3 complex 21 kDa subunit ARPC3 |
| 82 | d2bf5a1 |
|
not modelled |
5.5 |
19 |
Fold:Monooxygenase (hydroxylase) regulatory protein
Superfamily:Monooxygenase (hydroxylase) regulatory protein
Family:Monooxygenase (hydroxylase) regulatory protein |
| 83 | c2e5nA_ |
|
not modelled |
5.5 |
18 |
PDB header:transcription
Chain: A: PDB Molecule:rna polymerase ii elongation factor ell2;
PDBTitle: solution structure of the ell_n2 domain of target of rna2 polymerase ii elongation factor ell2
|
| 84 | c1bmv2_ |
|
not modelled |
5.4 |
21 |
PDB header:virus/rna
Chain: 2: PDB Molecule:protein (icosahedral virus - b and c domain);
PDBTitle: protein-rna interactions in an icosahedral virus at 3.02 angstroms resolution
|
| 85 | c1es6A_ |
|
not modelled |
5.4 |
43 |
PDB header:viral protein
Chain: A: PDB Molecule:matrix protein vp40;
PDBTitle: crystal structure of the matrix protein of ebola virus
|
| 86 | d1nexa2 |
|
not modelled |
5.3 |
20 |
Fold:POZ domain
Superfamily:POZ domain
Family:BTB/POZ domain |
| 87 | c2bfuL_ |
|
not modelled |
5.3 |
18 |
PDB header:virus
Chain: L: PDB Molecule:cowpea mosaic virus, large (l) subunit;
PDBTitle: x-ray structure of cpmv top component
|

