1 c2aexA_
100.0
48
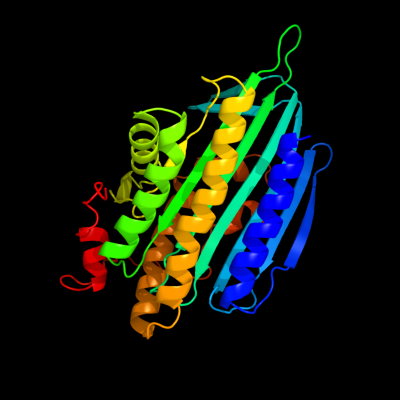
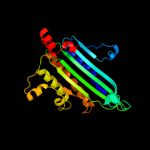
PDB header: oxidoreductaseChain: A: PDB Molecule: coproporphyrinogen iii oxidase, mitochondrial;PDBTitle: the 1.58a crystal structure of human coproporphyrinogen oxidase2 reveals the structural basis of hereditary coproporphyria
2 d1tkla_
100.0
47
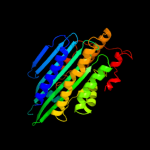
Fold: Coproporphyrinogen III oxidaseSuperfamily: Coproporphyrinogen III oxidaseFamily: Coproporphyrinogen III oxidase3 d1vjua_
100.0
66
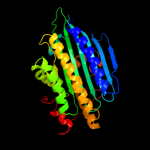
Fold: Coproporphyrinogen III oxidaseSuperfamily: Coproporphyrinogen III oxidaseFamily: Coproporphyrinogen III oxidase4 d1txna_
100.0
44
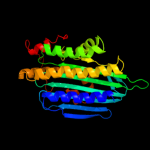
Fold: Coproporphyrinogen III oxidaseSuperfamily: Coproporphyrinogen III oxidaseFamily: Coproporphyrinogen III oxidase5 c3lm3A_
47.3
42
PDB header: unknown functionChain: A: PDB Molecule: uncharacterized protein;PDBTitle: crystal structure of a putative glycoside hydrolase/deacetylase2 (bdi_3119) from parabacteroides distasonis at 1.44 a resolution
6 d1ci4a_
35.3
18
Fold: SAM domain-likeSuperfamily: Barrier-to-autointegration factor, BAFFamily: Barrier-to-autointegration factor, BAF7 d1kkoa2
28.7
40
Fold: Enolase N-terminal domain-likeSuperfamily: Enolase N-terminal domain-likeFamily: Enolase N-terminal domain-like8 d1kcza2
28.3
50
Fold: Enolase N-terminal domain-likeSuperfamily: Enolase N-terminal domain-likeFamily: Enolase N-terminal domain-like9 c3fubA_
27.9
27
PDB header: hormoneChain: A: PDB Molecule: gdnf family receptor alpha-1;PDBTitle: crystal structure of gdnf-gfralpha1 complex
10 c2v5eA_
27.3
27
PDB header: receptor/glycoprotein complexChain: A: PDB Molecule: gdnf family receptor alpha-1;PDBTitle: the structure of the gdnf\:coreceptor complex\: insights2 into ret signalling and heparin binding.
11 d1y7pa2
26.9
26
Fold: Ferredoxin-likeSuperfamily: ACT-likeFamily: AF1403 N-terminal domain-like12 c2r7tA_
21.7
38
PDB header: transferase/rnaChain: A: PDB Molecule: rna-dependent rna polymerase;PDBTitle: crystal structure of rotavirus sa11 vp1/rna (ugugaacc)2 complex
13 c2r39A_
21.1
27
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: fixg-related protein;PDBTitle: crystal structure of fixg-related protein from vibrio parahaemolyticus
14 c1kkoB_
18.5
40
PDB header: lyaseChain: B: PDB Molecule: 3-methylaspartate ammonia-lyase;PDBTitle: crystal structure of citrobacter amalonaticus2 methylaspartate ammonia lyase
15 d1j2ga1
16.7
15
Fold: T-foldSuperfamily: Tetrahydrobiopterin biosynthesis enzymes-likeFamily: Urate oxidase (uricase)16 d1bdfa1
16.1
33
Fold: DCoH-likeSuperfamily: RBP11-like subunits of RNA polymeraseFamily: RNA polymerase alpha subunit dimerisation domain17 d1t9ka_
16.0
18
Fold: NagB/RpiA/CoA transferase-likeSuperfamily: NagB/RpiA/CoA transferase-likeFamily: IF2B-like18 d1gg4a1
15.7
21
Fold: MurD-like peptide ligases, peptide-binding domainSuperfamily: MurD-like peptide ligases, peptide-binding domainFamily: MurCDEF C-terminal domain19 c2pbyB_
14.9
71
PDB header: hydrolaseChain: B: PDB Molecule: glutaminase;PDBTitle: probable glutaminase from geobacillus kaustophilus hta426
20 d1yuaa1
13.1
47
Fold: Rubredoxin-likeSuperfamily: Zinc beta-ribbonFamily: Prokaryotic DNA topoisomerase I, a C-terminal fragment21 c3ccjU_
not modelled
13.0
29
PDB header: ribosomeChain: U: PDB Molecule: 50s ribosomal protein l24e;PDBTitle: structure of anisomycin resistant 50s ribosomal subunit: 23s rrna2 mutation c2534u
22 d1vb5a_
not modelled
12.8
22
Fold: NagB/RpiA/CoA transferase-likeSuperfamily: NagB/RpiA/CoA transferase-likeFamily: IF2B-like23 c2yvkA_
not modelled
12.7
16
PDB header: isomeraseChain: A: PDB Molecule: methylthioribose-1-phosphate isomerase;PDBTitle: crystal structure of 5-methylthioribose 1-phosphate2 isomerase product complex from bacillus subtilis
24 c3llxA_
not modelled
12.5
12
PDB header: isomeraseChain: A: PDB Molecule: predicted amino acid aldolase or racemase;PDBTitle: crystal structure of an ala racemase-like protein (il1761) from2 idiomarina loihiensis at 1.50 a resolution
25 d1vqou1
not modelled
12.3
29
Fold: Glucocorticoid receptor-like (DNA-binding domain)Superfamily: Glucocorticoid receptor-like (DNA-binding domain)Family: Ribosomal protein L24e26 d1auna_
not modelled
12.0
38
Fold: Osmotin, thaumatin-like proteinSuperfamily: Osmotin, thaumatin-like proteinFamily: Osmotin, thaumatin-like protein27 d2yzca1
not modelled
11.7
26
Fold: T-foldSuperfamily: Tetrahydrobiopterin biosynthesis enzymes-likeFamily: Urate oxidase (uricase)28 c2zkru_
not modelled
11.6
43
PDB header: ribosomal protein/rnaChain: U: PDB Molecule: rna expansion segment es41;PDBTitle: structure of a mammalian ribosomal 60s subunit within an2 80s complex obtained by docking homology models of the rna3 and proteins into an 8.7 a cryo-em map
29 d1wmxb_
not modelled
11.6
35
Fold: Galactose-binding domain-likeSuperfamily: Galactose-binding domain-likeFamily: Family 30 carbohydrate binding module, CBM30 (PKD repeat)30 c1w2wJ_
not modelled
11.5
22
PDB header: isomeraseChain: J: PDB Molecule: 5-methylthioribose-1-phosphate isomerase;PDBTitle: crystal structure of yeast ypr118w, a methylthioribose-1-2 phosphate isomerase related to regulatory eif2b subunits
31 c3uo9B_
not modelled
11.3
14
PDB header: hydrolase/hydrolase inhibitorChain: B: PDB Molecule: glutaminase kidney isoform, mitochondrial;PDBTitle: crystal structure of human gac in complex with glutamate and bptes
32 c2d35A_
not modelled
11.1
60
PDB header: cell cycleChain: A: PDB Molecule: cell division activator ceda;PDBTitle: solution structure of cell division reactivation factor,2 ceda
33 c1yuaA_
not modelled
10.9
47
PDB header: dna binding proteinChain: A: PDB Molecule: topoisomerase i;PDBTitle: c-terminal domain of escherichia coli topoisomerase i
34 c2bn8A_
not modelled
10.9
60
PDB header: cell cycle proteinChain: A: PDB Molecule: cell division activator ceda;PDBTitle: solution structure and interactions of the e.coli cell2 division activator protein ceda
35 c2yk5A_
not modelled
10.7
13
PDB header: transferaseChain: A: PDB Molecule: cmp-n-acetylneuraminate-beta-galactosamide-alpha-2,3-PDBTitle: structure of neisseria los-specific sialyltransferase (nst),2 in complex with cmp.
36 c3n2bD_
not modelled
10.5
17
PDB header: lyaseChain: D: PDB Molecule: diaminopimelate decarboxylase;PDBTitle: 1.8 angstrom resolution crystal structure of diaminopimelate2 decarboxylase (lysa) from vibrio cholerae.
37 d2fkla1
not modelled
10.0
33
Fold: Dodecin subunit-likeSuperfamily: Amyloid beta a4 protein copper binding domain (domain 2)Family: Amyloid beta a4 protein copper binding domain (domain 2)38 c3ecsD_
not modelled
9.9
24
PDB header: translationChain: D: PDB Molecule: translation initiation factor eif-2b subunitPDBTitle: crystal structure of human eif2b alpha
39 c4a1eT_
not modelled
9.6
29
PDB header: ribosomeChain: T: PDB Molecule: rpl24;PDBTitle: t.thermophila 60s ribosomal subunit in complex with2 initiation factor 6. this file contains 5s rrna, 5.8s rrna3 and proteins of molecule 1
40 d1ccwb_
not modelled
9.5
23
Fold: TIM beta/alpha-barrelSuperfamily: Cobalamin (vitamin B12)-dependent enzymesFamily: Glutamate mutase, large subunit41 c1u2zC_
not modelled
9.4
14
PDB header: transferaseChain: C: PDB Molecule: histone-lysine n-methyltransferase, h3 lysine-79PDBTitle: crystal structure of histone k79 methyltransferase dot1p2 from yeast
42 c2ebsB_
not modelled
9.2
21
PDB header: hydrolaseChain: B: PDB Molecule: oligoxyloglucan reducing end-specificPDBTitle: crystal structure anaalysis of oligoxyloglucan reducing-end-2 specific cellobiohydrolase (oxg-rcbh) d465n mutant3 complexed with a xyloglucan heptasaccharide
43 c3f42A_
not modelled
9.2
24
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: protein hp0035;PDBTitle: crystal structure of uncharacterized protein hp0035 from helicobacter2 pylori
44 c3izcZ_
not modelled
9.1
29
PDB header: ribosomeChain: Z: PDB Molecule: 60s ribosomal protein rpl24 (l24e);PDBTitle: localization of the large subunit ribosomal proteins into a 6.1 a2 cryo-em map of saccharomyces cerevisiae translating 80s ribosome
45 d1e88a3
not modelled
9.1
38
Fold: FnI-like domainSuperfamily: FnI-like domainFamily: Fibronectin type I module46 d2piaa2
not modelled
9.1
18
Fold: Ferredoxin reductase-like, C-terminal NADP-linked domainSuperfamily: Ferredoxin reductase-like, C-terminal NADP-linked domainFamily: Aromatic dioxygenase reductase-like47 d1uc8a1
not modelled
8.8
14
Fold: PreATP-grasp domainSuperfamily: PreATP-grasp domainFamily: Lysine biosynthesis enzyme LysX, N-terminal domain48 d1tvca2
not modelled
8.8
22
Fold: Ferredoxin reductase-like, C-terminal NADP-linked domainSuperfamily: Ferredoxin reductase-like, C-terminal NADP-linked domainFamily: Aromatic dioxygenase reductase-like49 d1u5la_
not modelled
8.6
10
Fold: Prion-likeSuperfamily: Prion-likeFamily: Prion-like50 d1tz9a_
not modelled
8.3
13
Fold: TIM beta/alpha-barrelSuperfamily: Xylose isomerase-likeFamily: UxuA-like51 d1tzpa_
not modelled
8.2
35
Fold: Hedgehog/DD-peptidaseSuperfamily: Hedgehog/DD-peptidaseFamily: MepA-like52 d2a0ua1
not modelled
8.1
16
Fold: NagB/RpiA/CoA transferase-likeSuperfamily: NagB/RpiA/CoA transferase-likeFamily: IF2B-like53 d1gff1_
not modelled
7.9
67
Fold: Nucleoplasmin-like/VP (viral coat and capsid proteins)Superfamily: ssDNA virusesFamily: Microviridae-like VP54 d1qfja2
not modelled
7.9
14
Fold: Ferredoxin reductase-like, C-terminal NADP-linked domainSuperfamily: Ferredoxin reductase-like, C-terminal NADP-linked domainFamily: Reductases55 c3iz5Z_
not modelled
7.7
29
PDB header: ribosomeChain: Z: PDB Molecule: 60s ribosomal protein l24 (l24e);PDBTitle: localization of the large subunit ribosomal proteins into a 5.5 a2 cryo-em map of triticum aestivum translating 80s ribosome
56 d1pzra_
not modelled
7.7
86
Fold: HLH-likeSuperfamily: Docking domain B of the erythromycin polyketide synthase (DEBS)Family: Docking domain B of the erythromycin polyketide synthase (DEBS)57 d1ulva3
not modelled
7.7
40
Fold: Immunoglobulin-like beta-sandwichSuperfamily: CBD9-likeFamily: Glucodextranase, domain C58 d2ibaa1
not modelled
7.5
16
Fold: T-foldSuperfamily: Tetrahydrobiopterin biosynthesis enzymes-likeFamily: Urate oxidase (uricase)59 d3be7a1
not modelled
7.4
27
Fold: Composite domain of metallo-dependent hydrolasesSuperfamily: Composite domain of metallo-dependent hydrolasesFamily: Zn-dependent arginine carboxypeptidase-like60 c3cxjB_
not modelled
7.4
15
PDB header: structural genomics, unknown functionChain: B: PDB Molecule: uncharacterized protein;PDBTitle: crystal structure of an uncharacterized protein from2 methanothermobacter thermautotrophicus
61 c2jv4A_
not modelled
7.4
25
PDB header: isomeraseChain: A: PDB Molecule: peptidyl-prolyl cis/trans isomerase;PDBTitle: structure characterisation of pina ww domain and comparison2 with other group iv ww domains, pin1 and ess1
62 c2xdyA_
not modelled
7.4
20
PDB header: rna binding proteinChain: A: PDB Molecule: post-transcriptional gene silencing protein qde-2;PDBTitle: crystal structure of the n. crassa qde-2 ago mid domain
63 d2d6fc2
not modelled
7.4
44
Fold: DCoH-likeSuperfamily: GAD domain-likeFamily: GAD domain64 d2bi0a1
not modelled
7.3
46
Fold: Thioesterase/thiol ester dehydrase-isomeraseSuperfamily: Thioesterase/thiol ester dehydrase-isomeraseFamily: MaoC-like65 d1edta_
not modelled
7.3
30
Fold: TIM beta/alpha-barrelSuperfamily: (Trans)glycosidasesFamily: Type II chitinase66 d1ny722
not modelled
7.3
27
Fold: Nucleoplasmin-like/VP (viral coat and capsid proteins)Superfamily: Positive stranded ssRNA virusesFamily: Comoviridae-like VP67 d1pcva_
not modelled
7.3
38
Fold: Osmotin, thaumatin-like proteinSuperfamily: Osmotin, thaumatin-like proteinFamily: Osmotin, thaumatin-like protein68 d1ddga2
not modelled
7.2
25
Fold: Ferredoxin reductase-like, C-terminal NADP-linked domainSuperfamily: Ferredoxin reductase-like, C-terminal NADP-linked domainFamily: NADPH-cytochrome p450 reductase-like69 c2rhfA_
not modelled
7.2
13
PDB header: hydrolaseChain: A: PDB Molecule: dna helicase recq;PDBTitle: d. radiodurans recq hrdc domain 3
70 c3n8bB_
not modelled
7.1
14
PDB header: nucleic acid binding proteinChain: B: PDB Molecule: uncharacterized protein;PDBTitle: crystal structure of borrelia burgdorferi pur-alpha
71 c1i7wB_
not modelled
7.1
50
PDB header: cell adhesionChain: B: PDB Molecule: epithelial-cadherin;PDBTitle: beta-catenin/phosphorylated e-cadherin complex
72 d1e8ga1
not modelled
7.0
10
Fold: Ferredoxin-likeSuperfamily: FAD-linked oxidases, C-terminal domainFamily: Vanillyl-alcohol oxidase-like73 c3nzpA_
not modelled
7.0
22
PDB header: lyaseChain: A: PDB Molecule: arginine decarboxylase;PDBTitle: crystal structure of the biosynthetic arginine decarboxylase spea from2 campylobacter jejuni, northeast structural genomics consortium target3 br53
74 c3nm7C_
not modelled
7.0
14
PDB header: nucleic acid binding proteinChain: C: PDB Molecule: uncharacterized protein;PDBTitle: crystal structure of borrelia burgdorferi pur-alpha
75 d1pgl22
not modelled
6.9
36
Fold: Nucleoplasmin-like/VP (viral coat and capsid proteins)Superfamily: Positive stranded ssRNA virusesFamily: Comoviridae-like VP76 c2j3zA_
not modelled
6.9
18
PDB header: toxinChain: A: PDB Molecule: c2 toxin component i;PDBTitle: crystal structure of the enzymatic component c2-i of the2 c2-toxin from clostridium botulinum at ph 6.1
77 c3nzqB_
not modelled
6.8
41
PDB header: lyaseChain: B: PDB Molecule: biosynthetic arginine decarboxylase;PDBTitle: crystal structure of biosynthetic arginine decarboxylase adc (spea)2 from escherichia coli, northeast structural genomics consortium3 target er600
78 c3nm7D_
not modelled
6.7
14
PDB header: nucleic acid binding proteinChain: D: PDB Molecule: uncharacterized protein;PDBTitle: crystal structure of borrelia burgdorferi pur-alpha
79 c3n8bA_
not modelled
6.7
14
PDB header: nucleic acid binding proteinChain: A: PDB Molecule: uncharacterized protein;PDBTitle: crystal structure of borrelia burgdorferi pur-alpha
80 d1ehsa_
not modelled
6.6
19
Fold: Toxic hairpinSuperfamily: Heat-stable enterotoxin BFamily: Heat-stable enterotoxin B81 c3ik2A_
not modelled
6.5
6
PDB header: hydrolaseChain: A: PDB Molecule: endoglucanase a;PDBTitle: crystal structure of a glycoside hydrolase family 44 endoglucanase2 produced by clostridium acetobutylium atcc 824
82 c3aagA_
not modelled
6.3
44
PDB header: transferaseChain: A: PDB Molecule: general glycosylation pathway protein;PDBTitle: crystal structure of c. jejuni pglb c-terminal domain
83 c1tvcA_
not modelled
6.3
21
PDB header: oxidoreductaseChain: A: PDB Molecule: methane monooxygenase component c;PDBTitle: fad and nadh binding domain of methane monooxygenase2 reductase from methylococcus capsulatus (bath)
84 c2j10B_
not modelled
6.2
58
PDB header: transcriptionChain: B: PDB Molecule: cellular tumor antigen p53;PDBTitle: p53 tetramerization domain mutant t329f q331k
85 c2ahnA_
not modelled
6.2
38
PDB header: allergenChain: A: PDB Molecule: thaumatin-like protein;PDBTitle: high resolution structure of a cherry allergen pru av 2
86 d1qt1a_
not modelled
6.2
19
Fold: TIM beta/alpha-barrelSuperfamily: Xylose isomerase-likeFamily: Xylose isomerase87 c2j10D_
not modelled
6.1
58
PDB header: transcriptionChain: D: PDB Molecule: cellular tumor antigen p53;PDBTitle: p53 tetramerization domain mutant t329f q331k
88 c2j10A_
not modelled
6.1
58
PDB header: transcriptionChain: A: PDB Molecule: cellular tumor antigen p53;PDBTitle: p53 tetramerization domain mutant t329f q331k
89 c3a11D_
not modelled
6.1
20
PDB header: isomeraseChain: D: PDB Molecule: translation initiation factor eif-2b, deltaPDBTitle: crystal structure of ribose-1,5-bisphosphate isomerase from2 thermococcus kodakaraensis kod1
90 c3ic8D_
not modelled
6.0
19
PDB header: structural genomics, unknown functionChain: D: PDB Molecule: uncharacterized gst-like proteinprotein;PDBTitle: the crystal structure of a gst-like protein from pseudomonas syringae2 to 2.4a
91 d1rqwa_
not modelled
5.9
38
Fold: Osmotin, thaumatin-like proteinSuperfamily: Osmotin, thaumatin-like proteinFamily: Osmotin, thaumatin-like protein92 c2k5dA_
not modelled
5.8
23
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: uncharacterized protein sag0934;PDBTitle: solution nmr structure of sag0934 from streptococcus2 agalactiae. northeast structural genomics target sar32[1-3 108].
93 d2aama1
not modelled
5.8
12
Fold: TIM beta/alpha-barrelSuperfamily: (Trans)glycosidasesFamily: TM1410-like94 c2aamA_
not modelled
5.8
12
PDB header: hydrolaseChain: A: PDB Molecule: hypothetical protein tm1410;PDBTitle: crystal structure of a putative glycosidase (tm1410) from thermotoga2 maritima at 2.20 a resolution
95 d1ry7a_
not modelled
5.8
16
Fold: beta-TrefoilSuperfamily: CytokineFamily: Fibroblast growth factors (FGF)96 d1qqha_
not modelled
5.8
8
Fold: E2 regulatory, transactivation domainSuperfamily: E2 regulatory, transactivation domainFamily: E2 regulatory, transactivation domain97 c3mt1B_
not modelled
5.7
15
PDB header: lyaseChain: B: PDB Molecule: putative carboxynorspermidine decarboxylase protein;PDBTitle: crystal structure of putative carboxynorspermidine decarboxylase2 protein from sinorhizobium meliloti
98 c1s1iS_
not modelled
5.6
29
PDB header: ribosomeChain: S: PDB Molecule: 60s ribosomal protein l24-a;PDBTitle: structure of the ribosomal 80s-eef2-sordarin complex from2 yeast obtained by docking atomic models for rna and protein3 components into a 11.7 a cryo-em map. this file, 1s1i,4 contains 60s subunit. the 40s ribosomal subunit is in file5 1s1h.
99 d1t3la1
not modelled
5.6
24
Fold: SH3-like barrelSuperfamily: SH3-domainFamily: SH3-domain