| 1 |
|
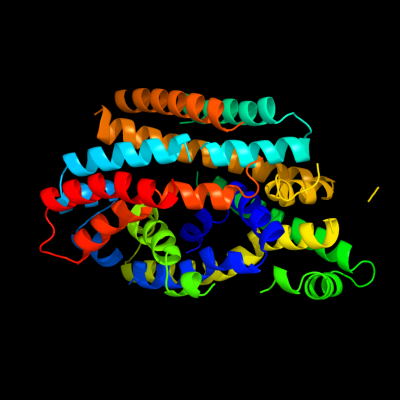
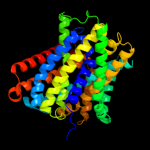
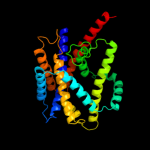
PDB 2xq2 chain A
Region: 25 - 400
Aligned: 373
Modelled: 373
Confidence: 99.9%
Identity: 12%
PDB header:transport protein
Chain: A: PDB Molecule:sodium/glucose cotransporter;
PDBTitle: structure of the k294a mutant of vsglt
Phyre2
| 2 |
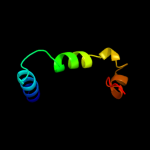
|
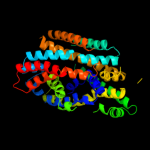
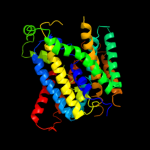
PDB 3dh4 chain A
Region: 25 - 398
Aligned: 371
Modelled: 374
Confidence: 99.9%
Identity: 12%
PDB header:transport protein
Chain: A: PDB Molecule:sodium/glucose cotransporter;
PDBTitle: crystal structure of sodium/sugar symporter with bound galactose from2 vibrio parahaemolyticus
Phyre2
| 3 |
|
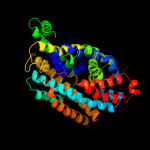
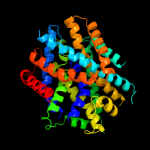
PDB 2jln chain A
Region: 3 - 402
Aligned: 397
Modelled: 397
Confidence: 99.7%
Identity: 10%
PDB header:membrane protein
Chain: A: PDB Molecule:mhp1;
PDBTitle: structure of mhp1, a nucleobase-cation-symport-1 family2 transporter
Phyre2
| 4 |
|
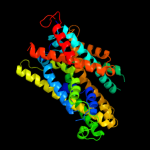
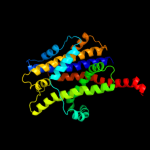
PDB 3gia chain A
Region: 15 - 411
Aligned: 386
Modelled: 386
Confidence: 99.4%
Identity: 10%
PDB header:transport protein
Chain: A: PDB Molecule:uncharacterized protein mj0609;
PDBTitle: crystal structure of apct transporter
Phyre2
| 5 |
|
PDB 3lrc chain C
Region: 15 - 400
Aligned: 355
Modelled: 355
Confidence: 98.7%
Identity: 13%
PDB header:transport protein
Chain: C: PDB Molecule:arginine/agmatine antiporter;
PDBTitle: structure of e. coli adic (p1)
Phyre2
| 6 |
|
PDB 2a65 chain A domain 1
Region: 26 - 407
Aligned: 381
Modelled: 381
Confidence: 98.1%
Identity: 13%
Fold: SNF-like
Superfamily: SNF-like
Family: SNF-like
Phyre2
| 7 |
|
PDB 2w8a chain C
Region: 89 - 387
Aligned: 299
Modelled: 299
Confidence: 92.3%
Identity: 11%
PDB header:membrane protein
Chain: C: PDB Molecule:glycine betaine transporter betp;
PDBTitle: crystal structure of the sodium-coupled glycine betaine2 symporter betp from corynebacterium glutamicum with bound3 substrate
Phyre2
| 8 |
|
PDB 3hfx chain A
Region: 89 - 381
Aligned: 282
Modelled: 293
Confidence: 84.4%
Identity: 12%
PDB header:transport protein
Chain: A: PDB Molecule:l-carnitine/gamma-butyrobetaine antiporter;
PDBTitle: crystal structure of carnitine transporter
Phyre2
| 9 |
|
PDB 2eyq chain A
Region: 6 - 52
Aligned: 47
Modelled: 47
Confidence: 6.8%
Identity: 23%
PDB header:hydrolase
Chain: A: PDB Molecule:transcription-repair coupling factor;
PDBTitle: crystal structure of escherichia coli transcription-repair2 coupling factor
Phyre2
| 10 |
|
PDB 1m3v chain A domain 2
Region: 15 - 24
Aligned: 10
Modelled: 10
Confidence: 6.7%
Identity: 30%
Fold: Glucocorticoid receptor-like (DNA-binding domain)
Superfamily: Glucocorticoid receptor-like (DNA-binding domain)
Family: LIM domain
Phyre2
| 11 |
|
PDB 2rlw chain A
Region: 8 - 30
Aligned: 23
Modelled: 23
Confidence: 6.5%
Identity: 22%
PDB header:toxin
Chain: A: PDB Molecule:plnf;
PDBTitle: three-dimensional structure of the two peptides that2 constitute the two-peptide bacteriocin plantaracin ef
Phyre2
| 12 |
|
PDB 2voy chain D
Region: 16 - 38
Aligned: 23
Modelled: 23
Confidence: 6.4%
Identity: 4%
PDB header:hydrolase
Chain: D: PDB Molecule:sarcoplasmic/endoplasmic reticulum calcium
PDBTitle: cryoem model of copa, the copper transporting atpase from2 archaeoglobus fulgidus
Phyre2
| 13 |
|
PDB 2hku chain A domain 2
Region: 36 - 52
Aligned: 17
Modelled: 17
Confidence: 6.4%
Identity: 41%
Fold: Tetracyclin repressor-like, C-terminal domain
Superfamily: Tetracyclin repressor-like, C-terminal domain
Family: Tetracyclin repressor-like, C-terminal domain
Phyre2
| 14 |
|
PDB 3mk7 chain F
Region: 53 - 110
Aligned: 58
Modelled: 58
Confidence: 6.2%
Identity: 10%
PDB header:oxidoreductase
Chain: F: PDB Molecule:cytochrome c oxidase, cbb3-type, subunit p;
PDBTitle: the structure of cbb3 cytochrome oxidase
Phyre2
| 15 |
|
PDB 2kwz chain A
Region: 16 - 31
Aligned: 16
Modelled: 16
Confidence: 6.0%
Identity: 38%
PDB header:viral protein
Chain: A: PDB Molecule:protease ns2-3;
PDBTitle: solution structure of ns2 [60-99]
Phyre2
| 16 |
|
PDB 3ajf chain A
Region: 358 - 381
Aligned: 24
Modelled: 24
Confidence: 5.4%
Identity: 8%
PDB header:viral protein
Chain: A: PDB Molecule:non-structural protein 3;
PDBTitle: structural insigths into dsrna binding and rna silencing suppression2 by ns3 protein of rice hoja blanca tenuivirus
Phyre2