| 1 | c1oj4B_
|
|
 |
100.0 |
98 |
PDB header:transferase
Chain: B: PDB Molecule:4-diphosphocytidyl-2-c-methyl-d-erythritol
PDBTitle: ternary complex of2 4-diphosphocytidyl-2-c-methyl-d-erythritol kinase
|
| 2 | c1uekA_
|
|
 |
100.0 |
36 |
PDB header:transferase
Chain: A: PDB Molecule:4-(cytidine 5'-diphospho)-2c-methyl-d-erythritol
PDBTitle: crystal structure of 4-(cytidine 5'-diphospho)-2c-methyl-d-2 erythritol kinase
|
| 3 | c2v34B_
|
|
 |
100.0 |
30 |
PDB header:transferase
Chain: B: PDB Molecule:4-diphosphocytidyl-2c-methyl-d-erythritol kinase;
PDBTitle: ispe in complex with cytidine and ligand
|
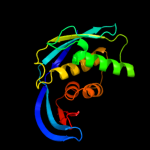
| 4 | d1oj4a1
|
|
 |
100.0 |
97 |
Fold:Ribosomal protein S5 domain 2-like
Superfamily:Ribosomal protein S5 domain 2-like
Family:GHMP Kinase, N-terminal domain |
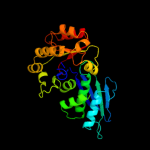
| 5 | c1fwlD_
|
|
 |
100.0 |
20 |
PDB header:transferase
Chain: D: PDB Molecule:homoserine kinase;
PDBTitle: crystal structure of homoserine kinase
|
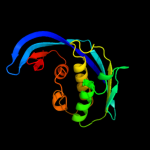
| 6 | d1ueka1
|
|
 |
100.0 |
44 |
Fold:Ribosomal protein S5 domain 2-like
Superfamily:Ribosomal protein S5 domain 2-like
Family:GHMP Kinase, N-terminal domain |
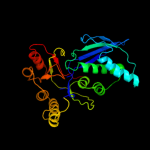
| 7 | c3hulA_
|
|
 |
100.0 |
17 |
PDB header:transferase
Chain: A: PDB Molecule:homoserine kinase;
PDBTitle: structure of putative homoserine kinase thrb from listeria2 monocytogenes
|
| 8 | c2gs8A_
|
|
 |
100.0 |
14 |
PDB header:lyase
Chain: A: PDB Molecule:mevalonate pyrophosphate decarboxylase;
PDBTitle: structure of mevalonate pyrophosphate decarboxylase from streptococcus2 pyogenes
|
| 9 | c2hfuB_
|
|
 |
100.0 |
15 |
PDB header:transferase
Chain: B: PDB Molecule:mevalonate kinase, putative;
PDBTitle: crystal structure of l. major mevalonate kinase in complex2 with r-mevalonate
|
| 10 | c1kkhA_
|
|
 |
100.0 |
18 |
PDB header:transferase
Chain: A: PDB Molecule:mevalonate kinase;
PDBTitle: crystal structure of the methanococcus jannaschii2 mevalonate kinase
|
| 11 | c2x7iA_
|
|
 |
100.0 |
16 |
PDB header:transferase
Chain: A: PDB Molecule:mevalonate kinase;
PDBTitle: crystal structure of mevalonate kinase from methicillin-2 resistant staphylococcus aureus mrsa252
|
| 12 | c2hk3A_
|
|
 |
100.0 |
15 |
PDB header:lyase
Chain: A: PDB Molecule:diphosphomevalonate decarboxylase;
PDBTitle: crystal structure of mevalonate diphosphate decarboxylase2 from staphylococcus aureus (orthorhombic form)
|
| 13 | c2oi2A_
|
|
 |
100.0 |
15 |
PDB header:transferase
Chain: A: PDB Molecule:mevalonate kinase;
PDBTitle: streptococcus pneumoniae mevalonate kinase in complex with2 diphosphomevalonate
|
| 14 | c2cz9A_
|
|
 |
100.0 |
22 |
PDB header:transferase
Chain: A: PDB Molecule:probable galactokinase;
PDBTitle: crystal structure of galactokinase from pyrococcus horikoshi
|
| 15 | c3k17A_
|
|
 |
100.0 |
15 |
PDB header:transferase
Chain: A: PDB Molecule:lin0012 protein;
PDBTitle: crystal structure of a lin0012 protein from listeria innocua
|
| 16 | c3ltoB_
|
|
 |
100.0 |
16 |
PDB header:lyase
Chain: B: PDB Molecule:mevalonate diphosphate decarboxylase;
PDBTitle: crystal structure of a mevalonate diphosphate decarboxylase2 from legionella pneumophila
|
| 17 | c3k85B_
|
|
 |
100.0 |
16 |
PDB header:transferase
Chain: B: PDB Molecule:d-glycero-d-manno-heptose 1-phosphate kinase;
PDBTitle: crystal structure of a d-glycero-d-manno-heptose 1-phosphate2 kinase from bacteriodes thetaiotaomicron
|
| 18 | c2r42A_
|
|
 |
100.0 |
20 |
PDB header:transferase
Chain: A: PDB Molecule:mevalonate kinase;
PDBTitle: the biochemical and structural basis for feedback2 inhibition of mevalonate kinase and isoprenoid metabolism
|
| 19 | c1pieA_
|
|
 |
100.0 |
20 |
PDB header:transferase
Chain: A: PDB Molecule:galactokinase;
PDBTitle: crystal structure of lactococcus lactis galactokinase2 complexed with galactose
|
| 20 | c1k47F_
|
|
 |
100.0 |
16 |
PDB header:transferase
Chain: F: PDB Molecule:phosphomevalonate kinase;
PDBTitle: crystal structure of the streptococcus pneumoniae2 phosphomevalonate kinase (pmk)
|
| 21 | c1wuuA_ |
|
not modelled |
100.0 |
21 |
PDB header:transferase
Chain: A: PDB Molecule:galactokinase;
PDBTitle: crystal structure of human galactokinase complexed with2 mgamppnp and galactose
|
| 22 | c2a2cA_ |
|
not modelled |
100.0 |
22 |
PDB header:transferase
Chain: A: PDB Molecule:n-acetylgalactosamine kinase;
PDBTitle: x-ray structure of human n-acetyl galactosamine kinase2 complexed with mg-adp and n-acetyl galactosamine 1-3 phosphate
|
| 23 | c2aj4B_ |
|
not modelled |
100.0 |
23 |
PDB header:transferase
Chain: B: PDB Molecule:galactokinase;
PDBTitle: crystal structure of saccharomyces cerevisiae galactokinase2 in complex with galactose and mg:amppnp
|
| 24 | d1oj4a2 |
|
not modelled |
100.0 |
98 |
Fold:Ferredoxin-like
Superfamily:GHMP Kinase, C-terminal domain
Family:4-(cytidine 5'-diphospho)-2C-methyl-D-erythritol kinase IspE |
| 25 | d1h72c1 |
|
not modelled |
100.0 |
20 |
Fold:Ribosomal protein S5 domain 2-like
Superfamily:Ribosomal protein S5 domain 2-like
Family:GHMP Kinase, N-terminal domain |
| 26 | d1kkha1 |
|
not modelled |
99.9 |
20 |
Fold:Ribosomal protein S5 domain 2-like
Superfamily:Ribosomal protein S5 domain 2-like
Family:GHMP Kinase, N-terminal domain |
| 27 | c2hkeB_ |
|
not modelled |
99.9 |
18 |
PDB header:lyase
Chain: B: PDB Molecule:diphosphomevalonate decarboxylase, putative;
PDBTitle: mevalonate diphosphate decarboxylase from trypanosoma brucei
|
| 28 | d1fi4a1 |
|
not modelled |
99.9 |
17 |
Fold:Ribosomal protein S5 domain 2-like
Superfamily:Ribosomal protein S5 domain 2-like
Family:GHMP Kinase, N-terminal domain |
| 29 | d1piea1 |
|
not modelled |
99.9 |
24 |
Fold:Ribosomal protein S5 domain 2-like
Superfamily:Ribosomal protein S5 domain 2-like
Family:GHMP Kinase, N-terminal domain |
| 30 | d1k47a1 |
|
not modelled |
99.9 |
18 |
Fold:Ribosomal protein S5 domain 2-like
Superfamily:Ribosomal protein S5 domain 2-like
Family:GHMP Kinase, N-terminal domain |
| 31 | d1wuua1 |
|
not modelled |
99.9 |
23 |
Fold:Ribosomal protein S5 domain 2-like
Superfamily:Ribosomal protein S5 domain 2-like
Family:GHMP Kinase, N-terminal domain |
| 32 | d1ueka2 |
|
not modelled |
99.9 |
24 |
Fold:Ferredoxin-like
Superfamily:GHMP Kinase, C-terminal domain
Family:4-(cytidine 5'-diphospho)-2C-methyl-D-erythritol kinase IspE |
| 33 | d1s4ea1 |
|
not modelled |
99.8 |
22 |
Fold:Ribosomal protein S5 domain 2-like
Superfamily:Ribosomal protein S5 domain 2-like
Family:GHMP Kinase, N-terminal domain |
| 34 | d1kvka1 |
|
not modelled |
99.8 |
25 |
Fold:Ribosomal protein S5 domain 2-like
Superfamily:Ribosomal protein S5 domain 2-like
Family:GHMP Kinase, N-terminal domain |
| 35 | d1h72c2 |
|
not modelled |
99.7 |
14 |
Fold:Ferredoxin-like
Superfamily:GHMP Kinase, C-terminal domain
Family:Homoserine kinase |
| 36 | c1fi4A_ |
|
not modelled |
99.5 |
15 |
PDB header:lyase
Chain: A: PDB Molecule:mevalonate 5-diphosphate decarboxylase;
PDBTitle: the x-ray crystal structure of mevalonate 5-diphosphate decarboxylase2 at 2.3 angstrom resolution.
|
| 37 | c3f0nB_ |
|
not modelled |
99.1 |
15 |
PDB header:lyase
Chain: B: PDB Molecule:mevalonate pyrophosphate decarboxylase;
PDBTitle: mus musculus mevalonate pyrophosphate decarboxylase
|
| 38 | d1kvka2 |
|
not modelled |
98.6 |
14 |
Fold:Ferredoxin-like
Superfamily:GHMP Kinase, C-terminal domain
Family:Mevalonate kinase |
| 39 | d1k47a2 |
|
not modelled |
98.6 |
12 |
Fold:Ferredoxin-like
Superfamily:GHMP Kinase, C-terminal domain
Family:Phosphomevalonate kinase (PMK) |
| 40 | d1kkha2 |
|
not modelled |
98.4 |
17 |
Fold:Ferredoxin-like
Superfamily:GHMP Kinase, C-terminal domain
Family:Mevalonate kinase |
| 41 | d1s4ea2 |
|
not modelled |
97.8 |
21 |
Fold:Ferredoxin-like
Superfamily:GHMP Kinase, C-terminal domain
Family:Galactokinase |
| 42 | d1wuua2 |
|
not modelled |
97.5 |
18 |
Fold:Ferredoxin-like
Superfamily:GHMP Kinase, C-terminal domain
Family:Galactokinase |
| 43 | d1piea2 |
|
not modelled |
97.4 |
15 |
Fold:Ferredoxin-like
Superfamily:GHMP Kinase, C-terminal domain
Family:Galactokinase |
| 44 | c3mnfA_ |
|
not modelled |
20.3 |
21 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:pac2 family protein;
PDBTitle: crystal structure of pac2 family protein from streptomyces avermitilis2 ma
|
| 45 | d1xxaa_ |
|
not modelled |
18.5 |
13 |
Fold:DCoH-like
Superfamily:C-terminal domain of arginine repressor
Family:C-terminal domain of arginine repressor |
| 46 | d1b4ba_ |
|
not modelled |
15.8 |
17 |
Fold:DCoH-like
Superfamily:C-terminal domain of arginine repressor
Family:C-terminal domain of arginine repressor |
| 47 | c3gaaB_ |
|
not modelled |
15.1 |
21 |
PDB header:structural genomics, unknown function
Chain: B: PDB Molecule:uncharacterized protein ta1441;
PDBTitle: the crystal structure of the protein with unknown function from2 thermoplasma acidophilum
|
| 48 | d2p90a1 |
|
not modelled |
14.4 |
10 |
Fold:Phosphorylase/hydrolase-like
Superfamily:Cgl1923-like
Family:Cgl1923-like |
| 49 | c2p90B_ |
|
not modelled |
13.4 |
10 |
PDB header:structural genomics, unknown function
Chain: B: PDB Molecule:hypothetical protein cgl1923;
PDBTitle: the crystal structure of a protein of unknown function from2 corynebacterium glutamicum atcc 13032
|
| 50 | c3cagF_ |
|
not modelled |
12.6 |
11 |
PDB header:dna binding protein
Chain: F: PDB Molecule:arginine repressor;
PDBTitle: crystal structure of the oligomerization domain hexamer of the2 arginine repressor protein from mycobacterium tuberculosis in complex3 with 9 arginines.
|
| 51 | c3fuxB_ |
|
not modelled |
11.2 |
21 |
PDB header:transferase
Chain: B: PDB Molecule:dimethyladenosine transferase;
PDBTitle: t. thermophilus 16s rrna a1518 and a1519 methyltransferase (ksga) in2 complex with 5'-methylthioadenosine in space group p212121
|
| 52 | c2wamB_ |
|
not modelled |
10.9 |
21 |
PDB header:unknown function
Chain: B: PDB Molecule:conserved hypothetical alanine and leucine rich
PDBTitle: crystal structure of mycobacterium tuberculosis unknown2 function protein rv2714
|
| 53 | d2p5ma1 |
|
not modelled |
10.9 |
13 |
Fold:DCoH-like
Superfamily:C-terminal domain of arginine repressor
Family:C-terminal domain of arginine repressor |
| 54 | c3e0dA_ |
|
not modelled |
10.7 |
17 |
PDB header:transferase/dna
Chain: A: PDB Molecule:dna polymerase iii subunit alpha;
PDBTitle: insights into the replisome from the crystral structure of2 the ternary complex of the eubacterial dna polymerase iii3 alpha-subunit
|
| 55 | d1yrxa1 |
|
not modelled |
10.2 |
11 |
Fold:Ferredoxin-like
Superfamily:Acylphosphatase/BLUF domain-like
Family:BLUF domain |
| 56 | d1a6qa1 |
|
not modelled |
8.5 |
35 |
Fold:Another 3-helical bundle
Superfamily:Protein serine/threonine phosphatase 2C, C-terminal domain
Family:Protein serine/threonine phosphatase 2C, C-terminal domain |
| 57 | d2byca1 |
|
not modelled |
8.5 |
10 |
Fold:Ferredoxin-like
Superfamily:Acylphosphatase/BLUF domain-like
Family:BLUF domain |
| 58 | c2hnhA_ |
|
not modelled |
8.3 |
25 |
PDB header:transferase
Chain: A: PDB Molecule:dna polymerase iii alpha subunit;
PDBTitle: crystal structure of the catalytic alpha subunit of e. coli2 replicative dna polymerase iii
|
| 59 | c2diuA_ |
|
not modelled |
8.1 |
13 |
PDB header:rna binding protein
Chain: A: PDB Molecule:kiaa0430 protein;
PDBTitle: solution structure of the rrm domain of kiaa0430 protein
|
| 60 | c3m05A_ |
|
not modelled |
8.1 |
18 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:uncharacterized protein pepe_1480;
PDBTitle: the crystal structure of a functionally unknown protein2 pepe_1480 from pediococcus pentosaceus atcc 25745
|
| 61 | d1o51a_ |
|
not modelled |
8.1 |
17 |
Fold:Ferredoxin-like
Superfamily:GlnB-like
Family:DUF190/COG1993 |
| 62 | c2hfnJ_ |
|
not modelled |
7.8 |
13 |
PDB header:electron transport
Chain: J: PDB Molecule:synechocystis photoreceptor (slr1694);
PDBTitle: crystal structures of the synechocystis photoreceptor slr1694 reveal2 distinct structural states related to signaling
|
| 63 | d1mwqa_ |
|
not modelled |
6.2 |
24 |
Fold:Ferredoxin-like
Superfamily:Dimeric alpha+beta barrel
Family:YciI-like |
| 64 | c3v4gA_ |
|
not modelled |
5.8 |
13 |
PDB header:dna binding protein
Chain: A: PDB Molecule:arginine repressor;
PDBTitle: 1.60 angstrom resolution crystal structure of an arginine repressor2 from vibrio vulnificus cmcp6
|
| 65 | d1vkra_ |
|
not modelled |
5.6 |
14 |
Fold:Phosphotyrosine protein phosphatases I-like
Superfamily:PTS system IIB component-like
Family:PTS system, Lactose/Cellobiose specific IIB subunit |
| 66 | c1vkrA_ |
|
not modelled |
5.6 |
14 |
PDB header:transferase
Chain: A: PDB Molecule:mannitol-specific pts system enzyme iiabc components;
PDBTitle: structure of iib domain of the mannitol-specific permease enzyme ii
|
| 67 | d8ohma2 |
|
not modelled |
5.5 |
14 |
Fold:P-loop containing nucleoside triphosphate hydrolases
Superfamily:P-loop containing nucleoside triphosphate hydrolases
Family:RNA helicase |
| 68 | d2gdwa1 |
|
not modelled |
5.4 |
26 |
Fold:Acyl carrier protein-like
Superfamily:ACP-like
Family:Peptidyl carrier domain |