| 1 |
|
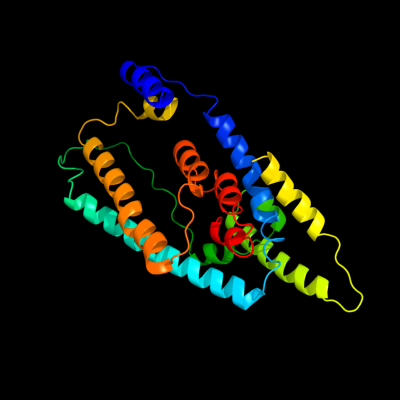
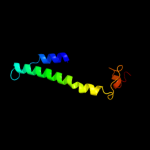
PDB 2nww chain A domain 1
Region: 39 - 376
Aligned: 256
Modelled: 256
Confidence: 93.7%
Identity: 17%
Fold: Proton glutamate symport protein
Superfamily: Proton glutamate symport protein
Family: Proton glutamate symport protein
Phyre2
| 2 |
|
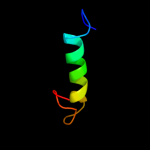
PDB 1zcd chain A
Region: 11 - 175
Aligned: 157
Modelled: 165
Confidence: 28.7%
Identity: 12%
PDB header:membrane protein
Chain: A: PDB Molecule:na(+)/h(+) antiporter 1;
PDBTitle: crystal structure of the na+/h+ antiporter nhaa
Phyre2
| 3 |
|
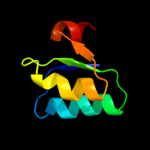
PDB 1e9y chain B domain 2
Region: 335 - 365
Aligned: 30
Modelled: 31
Confidence: 24.9%
Identity: 33%
Fold: TIM beta/alpha-barrel
Superfamily: Metallo-dependent hydrolases
Family: alpha-subunit of urease, catalytic domain
Phyre2
| 4 |
|
PDB 1ejx chain C domain 2
Region: 334 - 365
Aligned: 30
Modelled: 32
Confidence: 23.4%
Identity: 37%
Fold: TIM beta/alpha-barrel
Superfamily: Metallo-dependent hydrolases
Family: alpha-subunit of urease, catalytic domain
Phyre2
| 5 |
|
PDB 3b9y chain A
Region: 138 - 211
Aligned: 73
Modelled: 74
Confidence: 18.2%
Identity: 14%
PDB header:transport protein
Chain: A: PDB Molecule:ammonium transporter family rh-like protein;
PDBTitle: crystal structure of the nitrosomonas europaea rh protein
Phyre2
| 6 |
|
PDB 3chx chain F
Region: 11 - 52
Aligned: 42
Modelled: 42
Confidence: 8.7%
Identity: 19%
PDB header:membrane protein
Chain: F: PDB Molecule:pmoa;
PDBTitle: crystal structure of methylosinus trichosporium ob3b2 particulate methane monooxygenase (pmmo)
Phyre2
| 7 |
|
PDB 3bl2 chain A domain 1
Region: 364 - 395
Aligned: 32
Modelled: 32
Confidence: 8.4%
Identity: 19%
Fold: Toxins' membrane translocation domains
Superfamily: Bcl-2 inhibitors of programmed cell death
Family: Bcl-2 inhibitors of programmed cell death
Phyre2
| 8 |
|
PDB 1yew chain F
Region: 11 - 52
Aligned: 42
Modelled: 42
Confidence: 7.9%
Identity: 24%
PDB header:oxidoreductase, membrane protein
Chain: F: PDB Molecule:particulate methane monooxygenase, a subunit;
PDBTitle: crystal structure of particulate methane monooxygenase
Phyre2
| 9 |
|
PDB 2fna chain A domain 1
Region: 31 - 41
Aligned: 11
Modelled: 11
Confidence: 7.2%
Identity: 36%
Fold: DNA/RNA-binding 3-helical bundle
Superfamily: "Winged helix" DNA-binding domain
Family: Helicase DNA-binding domain
Phyre2
| 10 |
|
PDB 3gd8 chain A
Region: 19 - 125
Aligned: 107
Modelled: 107
Confidence: 7.1%
Identity: 19%
PDB header:membrane protein
Chain: A: PDB Molecule:aquaporin-4;
PDBTitle: crystal structure of human aquaporin 4 at 1.8 and its mechanism of2 conductance
Phyre2
| 11 |
|
PDB 2pv7 chain A domain 2
Region: 166 - 209
Aligned: 44
Modelled: 44
Confidence: 6.9%
Identity: 20%
Fold: NAD(P)-binding Rossmann-fold domains
Superfamily: NAD(P)-binding Rossmann-fold domains
Family: 6-phosphogluconate dehydrogenase-like, N-terminal domain
Phyre2
| 12 |
|
PDB 3cpi chain H
Region: 329 - 349
Aligned: 21
Modelled: 21
Confidence: 6.9%
Identity: 29%
PDB header:protein transport
Chain: H: PDB Molecule:rab gdp-dissociation inhibitor;
PDBTitle: crystal structure of yeast rab-gdi
Phyre2
| 13 |
|
PDB 1ivy chain A
Region: 134 - 153
Aligned: 18
Modelled: 20
Confidence: 6.4%
Identity: 39%
Fold: alpha/beta-Hydrolases
Superfamily: alpha/beta-Hydrolases
Family: Serine carboxypeptidase-like
Phyre2
| 14 |
|
PDB 3czc chain A
Region: 343 - 365
Aligned: 23
Modelled: 23
Confidence: 6.1%
Identity: 22%
PDB header:transferase
Chain: A: PDB Molecule:rmpb;
PDBTitle: the crystal structure of a putative pts iib(ptxb) from2 streptococcus mutans
Phyre2
| 15 |
|
PDB 2d57 chain A
Region: 19 - 112
Aligned: 94
Modelled: 94
Confidence: 6.1%
Identity: 19%
PDB header:transport protein
Chain: A: PDB Molecule:aquaporin-4;
PDBTitle: double layered 2d crystal structure of aquaporin-4 (aqp4m23) at 3.2 a2 resolution by electron crystallography
Phyre2
| 16 |
|
PDB 3hd6 chain A
Region: 138 - 211
Aligned: 74
Modelled: 74
Confidence: 5.9%
Identity: 12%
PDB header:membrane protein, transport protein
Chain: A: PDB Molecule:ammonium transporter rh type c;
PDBTitle: crystal structure of the human rhesus glycoprotein rhcg
Phyre2
| 17 |
|
PDB 1ohe chain A domain 2
Region: 342 - 370
Aligned: 27
Modelled: 29
Confidence: 5.5%
Identity: 33%
Fold: (Phosphotyrosine protein) phosphatases II
Superfamily: (Phosphotyrosine protein) phosphatases II
Family: Dual specificity phosphatase-like
Phyre2
| 18 |
|
PDB 3pdu chain F
Region: 166 - 220
Aligned: 55
Modelled: 55
Confidence: 5.3%
Identity: 16%
PDB header:oxidoreductase
Chain: F: PDB Molecule:3-hydroxyisobutyrate dehydrogenase family protein;
PDBTitle: crystal structure of gamma-hydroxybutyrate dehydrogenase from2 geobacter sulfurreducens in complex with nadp+
Phyre2