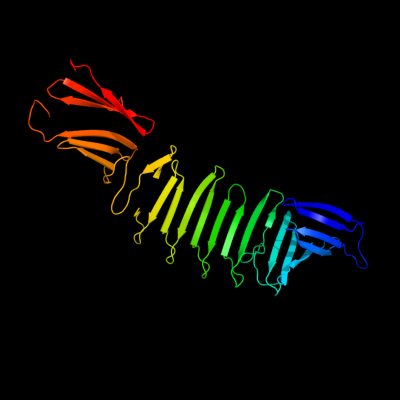
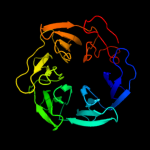
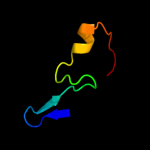
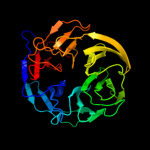
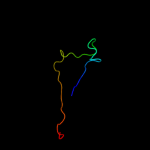
1 c2oy7A_
96.5
15
PDB header: membrane proteinChain: A: PDB Molecule: outer surface protein a;PDBTitle: the crystal structure of ospa mutant
2 d1rypi_
46.2
20
Fold: Ntn hydrolase-likeSuperfamily: N-terminal nucleophile aminohydrolases (Ntn hydrolases)Family: Proteasome subunits3 d1x3za1
46.0
18
Fold: Cysteine proteinasesSuperfamily: Cysteine proteinasesFamily: Transglutaminase core4 d1irua_
43.0
16
Fold: Ntn hydrolase-likeSuperfamily: N-terminal nucleophile aminohydrolases (Ntn hydrolases)Family: Proteasome subunits5 c3nqhA_
42.9
17
PDB header: hydrolaseChain: A: PDB Molecule: glycosyl hydrolase;PDBTitle: crystal structure of a glycosyl hydrolase (bt_2959) from bacteroides2 thetaiotaomicron vpi-5482 at 2.11 a resolution
6 c1j5qB_
37.5
25
PDB header: viral proteinChain: B: PDB Molecule: major capsid protein;PDBTitle: the structure and evolution of the major capsid protein of a large,2 lipid-containing, dna virus.
7 c1n7dA_
36.6
10
PDB header: lipid transportChain: A: PDB Molecule: low-density lipoprotein receptor;PDBTitle: extracellular domain of the ldl receptor
8 d1j2pa_
33.2
19
Fold: Ntn hydrolase-likeSuperfamily: N-terminal nucleophile aminohydrolases (Ntn hydrolases)Family: Proteasome subunits9 c1m4xC_
30.7
29
PDB header: virusChain: C: PDB Molecule: pbcv-1 virus capsid;PDBTitle: pbcv-1 virus capsid, quasi-atomic model
10 c3eswA_
30.5
18
PDB header: hydrolaseChain: A: PDB Molecule: peptide-n(4)-(n-acetyl-beta-glucosaminyl)asparaginePDBTitle: complex of yeast pngase with glcnac2-iac.
11 c2ivzD_
30.3
11
PDB header: protein transport/hydrolaseChain: D: PDB Molecule: protein tolb;PDBTitle: structure of tolb in complex with a peptide of the colicin2 e9 t-domain
12 d2f4ma1
28.5
26
Fold: Cysteine proteinasesSuperfamily: Cysteine proteinasesFamily: Transglutaminase core13 d1wc9a_
27.0
22
Fold: Ligand-binding domain in the NO signalling and Golgi transportSuperfamily: Ligand-binding domain in the NO signalling and Golgi transportFamily: TRAPP components14 c3e5zA_
26.8
15
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: putative gluconolactonase;PDBTitle: x-ray structure of the putative gluconolactonase in protein family2 pf08450. northeast structural genomics consortium target drr130.
15 d1m3ya2
26.1
29
Fold: Nucleoplasmin-like/VP (viral coat and capsid proteins)Superfamily: Group II dsDNA viruses VPFamily: Major capsid protein vp5416 c3mswA_
24.3
17
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: uncharacterized protein;PDBTitle: crystal structure of a protein with unknown function (bf3112) from2 bacteroides fragilis nctc 9343 at 1.90 a resolution
17 c2cfhA_
23.9
25
PDB header: transportChain: A: PDB Molecule: trafficking protein particle complex subunit 3;PDBTitle: structure of the bet3-tpc6b core of trapp
18 c1rz2A_
22.9
15
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: conserved hypothetical protein ba4783;PDBTitle: 1.6a crystal structure of the protein ba4783/q81l49 (similar to2 sortase b) from bacillus anthracis.
19 c3ifzA_
22.7
25
PDB header: isomeraseChain: A: PDB Molecule: dna gyrase subunit a;PDBTitle: crystal structure of the first part of the mycobacterium tuberculosis2 dna gyrase reaction core: the breakage and reunion domain at 2.7 a3 resolution
20 c2x12A_
22.4
27
PDB header: cell adhesionChain: A: PDB Molecule: fimbriae-associated protein fap1;PDBTitle: ph-induced modulation of streptococcus parasanguinis2 adhesion by fap1 fimbriae
21 c3rwxA_
not modelled
22.2
18
PDB header: transport proteinChain: A: PDB Molecule: hypothetical bacterial outer membrane protein;PDBTitle: crystal structure of a hypothetical bacterial outer membrane protein2 (bf2706) from bacteroides fragilis nctc 9343 at 2.40 a resolution
22 c2zcyM_
not modelled
20.2
16
PDB header: hydrolaseChain: M: PDB Molecule: proteasome component pre4;PDBTitle: yeast 20s proteasome:syringolin a-complex
23 d1ryp2_
not modelled
20.2
16
Fold: Ntn hydrolase-likeSuperfamily: N-terminal nucleophile aminohydrolases (Ntn hydrolases)Family: Proteasome subunits24 c1bqfA_
not modelled
20.1
70
PDB header: hormone/growth factorChain: A: PDB Molecule: protein (growth-blocking peptide);PDBTitle: growth-blocking peptide (gbp) from pseudaletia separata
25 d1rype_
not modelled
19.9
17
Fold: Ntn hydrolase-likeSuperfamily: N-terminal nucleophile aminohydrolases (Ntn hydrolases)Family: Proteasome subunits26 c2kijA_
not modelled
19.1
23
PDB header: hydrolaseChain: A: PDB Molecule: copper-transporting atpase 1;PDBTitle: solution structure of the actuator domain of the copper-2 transporting atpase atp7a
27 c2oryA_
not modelled
18.3
16
PDB header: hydrolaseChain: A: PDB Molecule: lipase;PDBTitle: crystal structure of m37 lipase
28 c3bmxB_
not modelled
17.8
38
PDB header: hydrolaseChain: B: PDB Molecule: uncharacterized lipoprotein ybbd;PDBTitle: beta-n-hexosaminidase (ybbd) from bacillus subtilis
29 d1ri9a_
not modelled
17.3
24
Fold: SH3-like barrelSuperfamily: SH3-domainFamily: SH3-domain30 c1ri9A_
not modelled
17.3
24
PDB header: signaling proteinChain: A: PDB Molecule: fyn-binding protein;PDBTitle: structure of a helically extended sh3 domain of the t cell2 adapter protein adap
31 d1pk6c_
not modelled
17.0
27
Fold: TNF-likeSuperfamily: TNF-likeFamily: TNF-like32 d1qwza_
not modelled
16.4
18
Fold: SortaseSuperfamily: SortaseFamily: Sortase33 c2ka3C_
not modelled
15.7
36
PDB header: structural proteinChain: C: PDB Molecule: emilin-1;PDBTitle: structure of emilin-1 c1q-like domain
34 d1rh1a1
not modelled
15.4
20
Fold: Cloacin translocation domainSuperfamily: Cloacin translocation domainFamily: Cloacin translocation domain35 c3nzwH_
not modelled
15.3
20
PDB header: hydrolase/hydrolase inhibitorChain: H: PDB Molecule: proteasome component pup1;PDBTitle: crystal structure of the yeast 20s proteasome in complex with 2b
36 d2oqza1
not modelled
15.1
15
Fold: SortaseSuperfamily: SortaseFamily: Sortase37 d1bcga_
not modelled
15.1
35
Fold: Knottins (small inhibitors, toxins, lectins)Superfamily: Scorpion toxin-likeFamily: Long-chain scorpion toxins38 c2gacA_
not modelled
15.1
16
PDB header: hydrolaseChain: A: PDB Molecule: glycosylasparaginase;PDBTitle: t152c mutant glycosylasparaginase from flavobacterium2 meningosepticum
39 c3h4pB_
not modelled
15.0
17
PDB header: hydrolaseChain: B: PDB Molecule: proteasome subunit alpha;PDBTitle: proteasome 20s core particle from methanocaldococcus2 jannaschii
40 d1bcoa1
not modelled
14.8
17
Fold: mu transposase, C-terminal domainSuperfamily: mu transposase, C-terminal domainFamily: mu transposase, C-terminal domain41 c3lk6A_
not modelled
14.7
38
PDB header: hydrolaseChain: A: PDB Molecule: lipoprotein ybbd;PDBTitle: beta-n-hexosaminidase n318d mutant (ybbd_n318d) from bacillus subtilis
42 d1pk6b_
not modelled
14.7
27
Fold: TNF-likeSuperfamily: TNF-likeFamily: TNF-like43 d1iruc_
not modelled
14.6
21
Fold: Ntn hydrolase-likeSuperfamily: N-terminal nucleophile aminohydrolases (Ntn hydrolases)Family: Proteasome subunits44 c3ilwA_
not modelled
14.4
45
PDB header: isomeraseChain: A: PDB Molecule: dna gyrase subunit a;PDBTitle: structure of dna gyrase subunit a n-terminal domain
45 d1xata_
not modelled
14.2
23
Fold: Single-stranded left-handed beta-helixSuperfamily: Trimeric LpxA-like enzymesFamily: Galactoside acetyltransferase-like46 c2kkyA_
not modelled
14.1
53
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: uncharacterized protein ecs2156;PDBTitle: solution structure of c-terminal domain of oxidized nleg2-3 (residue2 90-191) from pathogenic e. coli o157:h7. northeast structural3 genomics consortium and midwest center for structural genomics target4 et109a
47 d1rypc_
not modelled
13.8
22
Fold: Ntn hydrolase-likeSuperfamily: N-terminal nucleophile aminohydrolases (Ntn hydrolases)Family: Proteasome subunits48 c3ghpA_
not modelled
13.7
36
PDB header: structural proteinChain: A: PDB Molecule: cellulosomal scaffoldin adaptor protein b;PDBTitle: structure of the second type ii cohesin module from the2 adaptor scaa scaffoldin of acetivibrio cellulolyticus3 (including long c-terminal linker)
49 c1t3dB_
not modelled
13.7
32
PDB header: transferaseChain: B: PDB Molecule: serine acetyltransferase;PDBTitle: crystal structure of serine acetyltransferase from e.coli at 2.2a
50 d1c3ha_
not modelled
13.5
31
Fold: TNF-likeSuperfamily: TNF-likeFamily: TNF-like51 c2novD_
not modelled
13.4
19
PDB header: isomeraseChain: D: PDB Molecule: dna topoisomerase 4 subunit a;PDBTitle: breakage-reunion domain of s.pneumoniae topo iv: crystal2 structure of a gram-positive quinolone target
52 c2b59A_
not modelled
13.2
24
PDB header: hydrolase/structural proteinChain: A: PDB Molecule: cog1196: chromosome segregation atpases;PDBTitle: the type ii cohesin dockerin complex
53 d1rypj_
not modelled
13.0
15
Fold: Ntn hydrolase-likeSuperfamily: N-terminal nucleophile aminohydrolases (Ntn hydrolases)Family: Proteasome subunits54 c3vh0C_
not modelled
12.9
9
PDB header: protein binding/dnaChain: C: PDB Molecule: uncharacterized protein ynce;PDBTitle: crystal structure of e. coli ynce complexed with dna
55 c1zvuA_
not modelled
12.9
24
PDB header: isomeraseChain: A: PDB Molecule: topoisomerase iv subunit a;PDBTitle: structure of the full-length e. coli parc subunit
56 c2gtjA_
not modelled
12.7
18
PDB header: signaling proteinChain: A: PDB Molecule: fyn-binding protein;PDBTitle: reduced form of adap hsh3-n-domain
57 d1tyja1
not modelled
12.6
27
Fold: Common fold of diphtheria toxin/transcription factors/cytochrome fSuperfamily: Carbohydrate-binding domainFamily: Cellulose-binding domain family III58 d1zv9a1
not modelled
12.6
36
Fold: Common fold of diphtheria toxin/transcription factors/cytochrome fSuperfamily: Carbohydrate-binding domainFamily: Cellulose-binding domain family III59 d1irug_
not modelled
12.5
12
Fold: Ntn hydrolase-likeSuperfamily: N-terminal nucleophile aminohydrolases (Ntn hydrolases)Family: Proteasome subunits60 c2xetB_
not modelled
12.4
5
PDB header: transport proteinChain: B: PDB Molecule: f1 capsule-anchoring protein;PDBTitle: conserved hydrophobic clusters on the surface of the caf1a usher2 c-terminal domain are important for f1 antigen assembly
61 d1xaka_
not modelled
12.4
44
Fold: Immunoglobulin-like beta-sandwichSuperfamily: Accessory protein X4 (ORF8, ORF7a)Family: Accessory protein X4 (ORF8, ORF7a)62 c2w8bB_
not modelled
12.4
11
PDB header: protein transport/membrane proteinChain: B: PDB Molecule: protein tolb;PDBTitle: crystal structure of processed tolb in complex with pal
63 c2e5pA_
not modelled
12.3
23
PDB header: transcriptionChain: A: PDB Molecule: phd finger protein 1;PDBTitle: solution structure of the tudor domain of phd finger2 protein 1 (phf1 protein)
64 c3qx3B_
not modelled
12.2
50
PDB header: isomerase/dna/isomerase inhibitorChain: B: PDB Molecule: dna topoisomerase 2-beta;PDBTitle: human topoisomerase iibeta in complex with dna and etoposide
65 d1ng5a_
not modelled
12.1
18
Fold: SortaseSuperfamily: SortaseFamily: Sortase66 d1m7xa2
not modelled
12.0
13
Fold: Glycosyl hydrolase domainSuperfamily: Glycosyl hydrolase domainFamily: alpha-Amylases, C-terminal beta-sheet domain67 c2wl2B_
not modelled
11.8
22
PDB header: isomeraseChain: B: PDB Molecule: dna gyrase subunit a;PDBTitle: crystal structure of n-terminal domain of gyra with the2 antibiotic simocyclinone d8
68 c2iu9C_
not modelled
11.7
23
PDB header: transferaseChain: C: PDB Molecule: udp-3-o-[3-hydroxymyristoyl] glucosaminePDBTitle: chlamydia trachomatis lpxd with 100mm udpglcnac (complex ii)
69 d1ab4a_
not modelled
11.5
22
Fold: Type II DNA topoisomeraseSuperfamily: Type II DNA topoisomeraseFamily: Type II DNA topoisomerase70 c1p4vA_
not modelled
11.5
16
PDB header: hydrolaseChain: A: PDB Molecule: n(4)-(beta-n-acetylglucosaminyl)-l-asparaginasePDBTitle: crystal structure of the glycosylasparaginase precursor2 d151n mutant with glycine
71 c2wlgA_
not modelled
11.4
24
PDB header: transferaseChain: A: PDB Molecule: polysialic acid o-acetyltransferase;PDBTitle: crystallographic analysis of the polysialic acid o-2 acetyltransferase oatwy
72 d2jdqd1
not modelled
11.4
15
Fold: PB2 C-terminal domain-likeSuperfamily: PB2 C-terminal domain-likeFamily: PB2 C-terminal domain-like73 c2hc8A_
not modelled
11.4
29
PDB header: transport proteinChain: A: PDB Molecule: cation-transporting atpase, p-type;PDBTitle: structure of the a. fulgidus copa a-domain
74 c2xkjE_
not modelled
11.3
36
PDB header: isomeraseChain: E: PDB Molecule: topoisomerase iv;PDBTitle: crystal structure of catalytic core of a. baumannii topo iv2 (pare-parc fusion truncate)
75 c3fnkA_
not modelled
11.3
25
PDB header: structural proteinChain: A: PDB Molecule: cellulosomal scaffoldin adaptor protein b;PDBTitle: crystal structure of the second type ii cohesin module from2 the cellulosomal adaptor scaa scaffoldin of acetivibrio3 cellulolyticus
76 d1bjta_
not modelled
11.2
60
Fold: Type II DNA topoisomeraseSuperfamily: Type II DNA topoisomeraseFamily: Type II DNA topoisomerase77 c1bjtA_
not modelled
11.2
60
PDB header: topoisomeraseChain: A: PDB Molecule: topoisomerase ii;PDBTitle: topoisomerase ii residues 409-1201
78 d1t3da_
not modelled
11.2
32
Fold: Single-stranded left-handed beta-helixSuperfamily: Trimeric LpxA-like enzymesFamily: Serine acetyltransferase79 c2vtwF_
not modelled
11.2
27
PDB header: viral proteinChain: F: PDB Molecule: fiber protein 2;PDBTitle: structure of the c-terminal head domain of the fowl2 adenovirus type 1 short fibre
80 c1yo4A_
not modelled
11.1
47
PDB header: viral proteinChain: A: PDB Molecule: hypothetical protein x4;PDBTitle: solution structure of the sars coronavirus orf 7a coded x42 protein
81 c1hrlA_
not modelled
11.0
60
PDB header: toxinChain: A: PDB Molecule: paralytic peptide i;PDBTitle: structure of a paralytic peptide from an insect, manduca2 sexta
82 d1iruh_
not modelled
10.9
12
Fold: Ntn hydrolase-likeSuperfamily: N-terminal nucleophile aminohydrolases (Ntn hydrolases)Family: Proteasome subunits83 d1j2qh_
not modelled
10.8
25
Fold: Ntn hydrolase-likeSuperfamily: N-terminal nucleophile aminohydrolases (Ntn hydrolases)Family: Proteasome subunits84 d1iruk_
not modelled
10.8
22
Fold: Ntn hydrolase-likeSuperfamily: N-terminal nucleophile aminohydrolases (Ntn hydrolases)Family: Proteasome subunits85 d1pk6a_
not modelled
10.7
22
Fold: TNF-likeSuperfamily: TNF-likeFamily: TNF-like86 d1w7pd2
not modelled
10.6
25
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: Vacuolar sorting protein domain87 c3dr2A_
not modelled
10.5
14
PDB header: hydrolaseChain: A: PDB Molecule: exported gluconolactonase;PDBTitle: structural and functional analyses of xc5397 from2 xanthomonas campestris: a gluconolactonase important in3 glucose secondary metabolic pathways
88 d1x38a2
not modelled
10.5
25
Fold: Flavodoxin-likeSuperfamily: Beta-D-glucan exohydrolase, C-terminal domainFamily: Beta-D-glucan exohydrolase, C-terminal domain89 c2wh7A_
not modelled
10.4
30
PDB header: hydrolaseChain: A: PDB Molecule: hyaluronidase-phage associated;PDBTitle: the partial structure of a group a streptpcoccal phage-2 encoded tail fibre hyaluronate lyase hylp2
90 d1rypd_
not modelled
10.0
13
Fold: Ntn hydrolase-likeSuperfamily: N-terminal nucleophile aminohydrolases (Ntn hydrolases)Family: Proteasome subunits91 d1eaic_
not modelled
9.9
25
Fold: Serine protease inhibitorsSuperfamily: Serine protease inhibitorsFamily: ATI-like92 c2xk0A_
not modelled
9.9
27
PDB header: transcriptionChain: A: PDB Molecule: polycomb protein pcl;PDBTitle: solution structure of the tudor domain from drosophila2 polycomblike (pcl)
93 c2hh9A_
not modelled
9.9
28
PDB header: transferaseChain: A: PDB Molecule: thiamin pyrophosphokinase;PDBTitle: thiamin pyrophosphokinase from candida albicans
94 d1krra_
not modelled
9.8
21
Fold: Single-stranded left-handed beta-helixSuperfamily: Trimeric LpxA-like enzymesFamily: Galactoside acetyltransferase-like95 d1o5ua_
not modelled
9.8
11
Fold: Double-stranded beta-helixSuperfamily: RmlC-like cupinsFamily: Hypothetical protein TM111296 d1o91a_
not modelled
9.7
31
Fold: TNF-likeSuperfamily: TNF-likeFamily: TNF-like97 c1o91B_
not modelled
9.7
31
PDB header: collagenChain: B: PDB Molecule: collagen alpha 1(viii) chain;PDBTitle: crystal structure of a collagen viii nc1 domain trimer
98 d1jnpa_
not modelled
9.7
26
Fold: Oncogene productsSuperfamily: Oncogene productsFamily: Oncogene products99 c7apiB_
not modelled
9.6
6
PDB header: proteinase inhibitorChain: B: PDB Molecule: alpha 1-antitrypsin;PDBTitle: the s variant of human alpha1-antitrypsin, structure and implications2 for function and metabolism