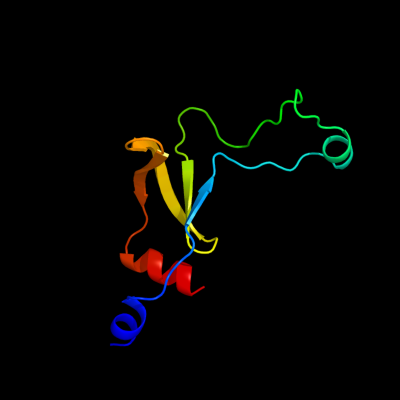
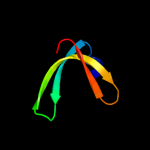
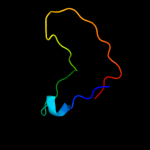
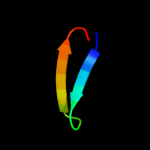
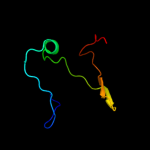
1 d1o6aa_
99.9
55
Fold: Surface presentation of antigens (SPOA)Superfamily: Surface presentation of antigens (SPOA)Family: Surface presentation of antigens (SPOA)2 d1o9ya_
99.8
27
Fold: Surface presentation of antigens (SPOA)Superfamily: Surface presentation of antigens (SPOA)Family: Surface presentation of antigens (SPOA)3 c2k5fA_
58.6
29
PDB header: metal transportChain: A: PDB Molecule: ferrous iron transport protein a;PDBTitle: solution nmr structure of feoa protein from chlorobium2 tepidum. northeast structural genomics consortium target3 ctr121
4 d2gcxa1
56.8
23
Fold: SH3-like barrelSuperfamily: C-terminal domain of transcriptional repressorsFamily: FeoA-like5 c2k5lA_
46.5
26
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: feoa;PDBTitle: solution nmr structure of protein feoa from clostridium2 thermocellum, northeast structural genomics consortium3 target cmr17
6 c2k4yA_
46.1
23
PDB header: metal transportChain: A: PDB Molecule: feoa-like protein;PDBTitle: nmr structure of feoa-like protein from clostridium2 acetobutylicum: northeast structural genomics consortium3 target car178
7 c3mhxB_
32.6
27
PDB header: metal transportChain: B: PDB Molecule: putative ferrous iron transport protein a;PDBTitle: crystal structure of stenotrophomonas maltophilia feoa complexed with2 zinc: a unique procaryotic sh3 domain protein possibly acting as a3 bacterial ferrous iron transport activating factor
8 c2k5iA_
32.1
19
PDB header: metal transportChain: A: PDB Molecule: iron transport protein;PDBTitle: solution structure of iron(ii) transport protein a from2 clostridium thermocellum , northeast structural genomics3 consortium (nesg) target vr131
9 c3gt2A_
29.5
17
PDB header: unknown functionChain: A: PDB Molecule: putative uncharacterized protein;PDBTitle: crystal structure of the p60 domain from m. avium2 paratuberculosis antigen map1272c
10 d3bwud1
26.5
21
Fold: FimD N-terminal domain-likeSuperfamily: FimD N-terminal domain-likeFamily: Usher N-domain11 d1m5q1_
26.3
13
Fold: Sm-like foldSuperfamily: Sm-like ribonucleoproteinsFamily: Sm motif of small nuclear ribonucleoproteins, SNRNP12 d1wmxb_
24.4
11
Fold: Galactose-binding domain-likeSuperfamily: Galactose-binding domain-likeFamily: Family 30 carbohydrate binding module, CBM30 (PKD repeat)13 d1zdva1
24.0
31
Fold: FimD N-terminal domain-likeSuperfamily: FimD N-terminal domain-likeFamily: Usher N-domain14 c3ddyA_
23.5
19
PDB header: luminescent proteinChain: A: PDB Molecule: lumazine protein;PDBTitle: structure of lumazine protein, an optical transponder of luminescent2 bacteria
15 d2h3ja1
23.3
17
Fold: SH3-like barrelSuperfamily: C-terminal domain of transcriptional repressorsFamily: FeoA-like16 c2glvA_
22.0
21
PDB header: lyaseChain: A: PDB Molecule: (3r)-hydroxymyristoyl-acyl carrier proteinPDBTitle: crystal structure of (3r)-hydroxyacyl-acyl carrier protein2 dehydratase(fabz) mutant(y100a) from helicobacter pylori
17 c2e63A_
20.8
23
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: kiaa1787 protein;PDBTitle: solution structure of the neuz domain in kiaa1787 protein
18 c2fg0B_
20.3
11
PDB header: hydrolaseChain: B: PDB Molecule: cog0791: cell wall-associated hydrolases (invasion-PDBTitle: crystal structure of a putative gamma-d-glutamyl-l-diamino acid2 endopeptidase (npun_r0659) from nostoc punctiforme pcc 73102 at 1.793 a resolution
19 d2evra2
20.1
11
Fold: Cysteine proteinasesSuperfamily: Cysteine proteinasesFamily: NlpC/P6020 c3e19D_
19.2
26
PDB header: transcription regulator, metal binding pChain: D: PDB Molecule: feoa;PDBTitle: crystal structure of iron uptake regulatory protein (feoa) solved by2 sulfur sad in a monoclinic space group
21 d2isba1
not modelled
19.0
33
Fold: The "swivelling" beta/beta/alpha domainSuperfamily: FumA C-terminal domain-likeFamily: FumA C-terminal domain-like22 c2kytA_
not modelled
17.9
7
PDB header: hydrolaseChain: A: PDB Molecule: group xvi phospholipase a2;PDBTitle: solution struture of the h-rev107 n-terminal domain
23 d1zdxa1
not modelled
17.8
26
Fold: FimD N-terminal domain-likeSuperfamily: FimD N-terminal domain-likeFamily: Usher N-domain24 c3pbiA_
not modelled
17.3
15
PDB header: hydrolaseChain: A: PDB Molecule: invasion protein;PDBTitle: structure of the peptidoglycan hydrolase ripb (rv1478) from2 mycobacterium tuberculosis at 1.6 resolution
25 c2xivA_
not modelled
15.9
19
PDB header: structural proteinChain: A: PDB Molecule: hypothetical invasion protein;PDBTitle: structure of rv1477, hypothetical invasion protein of2 mycobacterium tuberculosis
26 c1i8dB_
not modelled
15.3
18
PDB header: transferaseChain: B: PDB Molecule: riboflavin synthase;PDBTitle: crystal structure of riboflavin synthase
27 c3hruA_
not modelled
15.2
16
PDB header: transcriptionChain: A: PDB Molecule: metalloregulator scar;PDBTitle: crystal structure of scar with bound zn2+
28 c3cw1D_
not modelled
14.9
16
PDB header: splicingChain: D: PDB Molecule: small nuclear ribonucleoprotein sm d3;PDBTitle: crystal structure of human spliceosomal u1 snrnp
29 c2gl0A_
not modelled
14.9
22
PDB header: transferaseChain: A: PDB Molecule: conserved hypothetical protein;PDBTitle: structure of pae2307 in complex with adenosine
30 d1u1za_
not modelled
14.7
6
Fold: Thioesterase/thiol ester dehydrase-isomeraseSuperfamily: Thioesterase/thiol ester dehydrase-isomeraseFamily: FabZ-like31 d1vgga_
not modelled
14.6
16
Fold: Ta1353-likeSuperfamily: Ta1353-likeFamily: Ta1353-like32 c2d16B_
not modelled
14.3
16
PDB header: structural genomics, unknown functionChain: B: PDB Molecule: hypothetical protein ph1918;PDBTitle: crystal structure of ph1918 protein from pyrococcus horikoshii ot3
33 c2ekmC_
not modelled
14.2
23
PDB header: structural genomics, unknown functionChain: C: PDB Molecule: hypothetical protein st1511;PDBTitle: structure of st1219 protein from sulfolobus tokodaii
34 c3swnC_
not modelled
13.6
10
PDB header: rna binding proteinChain: C: PDB Molecule: u6 snrna-associated sm-like protein lsm7;PDBTitle: structure of the lsm657 complex: an assembly intermediate of the lsm12 7 and lsm2 8 rings
35 d1i4k1_
not modelled
13.2
14
Fold: Sm-like foldSuperfamily: Sm-like ribonucleoproteinsFamily: Sm motif of small nuclear ribonucleoproteins, SNRNP36 d1vyua1
not modelled
12.6
6
Fold: SH3-like barrelSuperfamily: SH3-domainFamily: SH3-domain37 c3cw15_
not modelled
12.0
19
PDB header: splicingChain: 5: PDB Molecule: small nuclear ribonucleoprotein g;PDB Fragment: residues 1-215;
PDBTitle: crystal structure of human spliceosomal u1 snrnp
38 d2je8a4
not modelled
11.8
15
Fold: Galactose-binding domain-likeSuperfamily: Galactose-binding domain-likeFamily: beta-Galactosidase/glucuronidase, N-terminal domain39 d1t0ha_
not modelled
11.7
12
Fold: SH3-like barrelSuperfamily: SH3-domainFamily: SH3-domain40 c2glwA_
not modelled
11.5
14
PDB header: transcriptionChain: A: PDB Molecule: 92aa long hypothetical protein;PDBTitle: the solution structure of phs018 from pyrococcus horikoshii
41 d4fiva_
not modelled
10.9
27
Fold: Acid proteasesSuperfamily: Acid proteasesFamily: Retroviral protease (retropepsin)42 c2pn0D_
not modelled
10.6
31
PDB header: transcriptionChain: D: PDB Molecule: prokaryotic transcription elongation factorPDBTitle: prokaryotic transcription elongation factor grea/greb from2 nitrosomonas europaea
43 d2as0a1
not modelled
10.6
23
Fold: PUA domain-likeSuperfamily: PUA domain-likeFamily: Hypothetical RNA methyltransferase domain (HRMD)44 d1n9ra_
not modelled
10.3
13
Fold: Sm-like foldSuperfamily: Sm-like ribonucleoproteinsFamily: Sm motif of small nuclear ribonucleoproteins, SNRNP45 c1b34A_
not modelled
10.2
16
PDB header: rna binding proteinChain: A: PDB Molecule: protein (small nuclear ribonucleoprotein sm d1);PDBTitle: crystal structure of the d1d2 sub-complex from the human snrnp core2 domain
46 d1b34a_
not modelled
10.2
16
Fold: Sm-like foldSuperfamily: Sm-like ribonucleoproteinsFamily: Sm motif of small nuclear ribonucleoproteins, SNRNP47 c3a35B_
not modelled
9.9
17
PDB header: luminescent proteinChain: B: PDB Molecule: lumazine protein;PDBTitle: crystal structure of lump complexed with riboflavin
48 d2axwa1
not modelled
9.7
19
Fold: Common fold of diphtheria toxin/transcription factors/cytochrome fSuperfamily: Bacterial adhesinsFamily: Pilus subunits49 d1d3ba_
not modelled
9.6
16
Fold: Sm-like foldSuperfamily: Sm-like ribonucleoproteinsFamily: Sm motif of small nuclear ribonucleoproteins, SNRNP50 c3d37A_
not modelled
9.5
21
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: tail protein, 43 kda;PDBTitle: the crystal structure of the tail protein from neisseria meningitidis2 mc58
51 d1t3la1
not modelled
9.2
12
Fold: SH3-like barrelSuperfamily: SH3-domainFamily: SH3-domain52 d2fwka1
not modelled
8.9
13
Fold: Sm-like foldSuperfamily: Sm-like ribonucleoproteinsFamily: Sm motif of small nuclear ribonucleoproteins, SNRNP53 d1eika_
not modelled
8.9
14
Fold: RPB5-like RNA polymerase subunitSuperfamily: RPB5-like RNA polymerase subunitFamily: RPB554 c3f3bA_
not modelled
8.9
21
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: phage-like element pbsx protein xkdh;PDBTitle: structure of the phage-like element pbsx protein xkdh from2 bacillus subtilus. northeast structural genomics3 consortium target sr352.
55 c1n9sH_
not modelled
8.6
13
PDB header: translationChain: H: PDB Molecule: small nuclear ribonucleoprotein f;PDBTitle: crystal structure of yeast smf in spacegroup p43212
56 c1t0jA_
not modelled
8.5
12
PDB header: signaling proteinChain: A: PDB Molecule: voltage-gated calcium channel subunit beta2a;PDBTitle: crystal structure of a complex between voltage-gated calcium channel2 beta2a subunit and a peptide of the alpha1c subunit
57 d1hmja_
not modelled
8.4
16
Fold: RPB5-like RNA polymerase subunitSuperfamily: RPB5-like RNA polymerase subunitFamily: RPB558 d1wmxa_
not modelled
8.1
8
Fold: Galactose-binding domain-likeSuperfamily: Galactose-binding domain-likeFamily: Family 30 carbohydrate binding module, CBM30 (PKD repeat)59 d1i8fa_
not modelled
8.1
11
Fold: Sm-like foldSuperfamily: Sm-like ribonucleoproteinsFamily: Sm motif of small nuclear ribonucleoproteins, SNRNP60 c3fcgB_
not modelled
8.1
27
PDB header: membrane protein, protein transportChain: B: PDB Molecule: f1 capsule-anchoring protein;PDBTitle: crystal structure analysis of the middle domain of the2 caf1a usher
61 c3swnA_
not modelled
7.7
18
PDB header: rna binding proteinChain: A: PDB Molecule: u6 snrna-associated sm-like protein lsm5;PDBTitle: structure of the lsm657 complex: an assembly intermediate of the lsm12 7 and lsm2 8 rings
62 d1h641_
not modelled
7.6
8
Fold: Sm-like foldSuperfamily: Sm-like ribonucleoproteinsFamily: Sm motif of small nuclear ribonucleoproteins, SNRNP63 c1fx7C_
not modelled
7.5
6
PDB header: signaling proteinChain: C: PDB Molecule: iron-dependent repressor ider;PDBTitle: crystal structure of the iron-dependent regulator (ider)2 from mycobacterium tuberculosis
64 c1wruA_
not modelled
7.4
14
PDB header: structural proteinChain: A: PDB Molecule: 43 kda tail protein;PDBTitle: structure of central hub elucidated by x-ray analysis of gene product2 44; baseplate component of bacteriophage mu
65 c3butA_
not modelled
7.3
38
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: uncharacterized protein af_0446;PDBTitle: crystal structure of protein af_0446 from archaeoglobus fulgidus
66 d1dzfa2
not modelled
7.3
25
Fold: RPB5-like RNA polymerase subunitSuperfamily: RPB5-like RNA polymerase subunitFamily: RPB567 d1d1na_
not modelled
7.1
26
Fold: Reductase/isomerase/elongation factor common domainSuperfamily: Translation proteinsFamily: Elongation factors68 d1jz8a3
not modelled
6.8
17
Fold: Galactose-binding domain-likeSuperfamily: Galactose-binding domain-likeFamily: beta-Galactosidase/glucuronidase, N-terminal domain69 c3h41A_
not modelled
6.7
14
PDB header: hydrolaseChain: A: PDB Molecule: nlp/p60 family protein;PDBTitle: crystal structure of a nlpc/p60 family protein (bce_2878) from2 bacillus cereus atcc 10987 at 1.79 a resolution
70 c3h0gE_
not modelled
6.6
25
PDB header: transcriptionChain: E: PDB Molecule: dna-directed rna polymerases i, ii, and iiiPDBTitle: rna polymerase ii from schizosaccharomyces pombe
71 c2fwkB_
not modelled
6.4
14
PDB header: dna binding proteinChain: B: PDB Molecule: u6 snrna-associated sm-like protein lsm5;PDBTitle: crystal structure of cryptosporidium parvum u6 snrna-associated sm-2 like protein lsm5
72 d2pw9a1
not modelled
6.4
25
Fold: Cytidine deaminase-likeSuperfamily: Cytidine deaminase-likeFamily: FdhD/NarQ73 c2fvnA_
not modelled
6.3
19
PDB header: cell adhesionChain: A: PDB Molecule: protein afad;PDBTitle: the fibrillar tip complex of the afa/dr adhesins from2 pathogen e. coli displays synergistic binding to 5 1 and v3 3 integrins
74 d1n9sc_
not modelled
6.3
13
Fold: Sm-like foldSuperfamily: Sm-like ribonucleoproteinsFamily: Sm motif of small nuclear ribonucleoproteins, SNRNP75 c2dlpA_
not modelled
6.2
27
PDB header: structural proteinChain: A: PDB Molecule: kiaa1783 protein;PDBTitle: solution structure of the sh3 domain of human kiaa17832 protein
76 c3cw1Z_
not modelled
6.2
22
PDB header: splicingChain: Z: PDB Molecule: small nuclear ribonucleoprotein f;PDBTitle: crystal structure of human spliceosomal u1 snrnp
77 d1mgqa_
not modelled
6.2
16
Fold: Sm-like foldSuperfamily: Sm-like ribonucleoproteinsFamily: Sm motif of small nuclear ribonucleoproteins, SNRNP78 c2yueA_
not modelled
6.0
7
PDB header: rna binding proteinChain: A: PDB Molecule: protein neuralized;PDBTitle: solution structure of the neuz (nhr) domain in neuralized2 from drosophila melanogaster
79 c3swnT_
not modelled
6.0
11
PDB header: rna binding proteinChain: T: PDB Molecule: u6 snrna-associated sm-like protein lsm6;PDBTitle: structure of the lsm657 complex: an assembly intermediate of the lsm12 7 and lsm2 8 rings
80 d1jbma_
not modelled
5.9
16
Fold: Sm-like foldSuperfamily: Sm-like ribonucleoproteinsFamily: Sm motif of small nuclear ribonucleoproteins, SNRNP81 d1o91a_
not modelled
5.7
17
Fold: TNF-likeSuperfamily: TNF-likeFamily: TNF-like82 c1o91B_
not modelled
5.7
17
PDB header: collagenChain: B: PDB Molecule: collagen alpha 1(viii) chain;PDBTitle: crystal structure of a collagen viii nc1 domain trimer
83 d1ljoa_
not modelled
5.6
13
Fold: Sm-like foldSuperfamily: Sm-like ribonucleoproteinsFamily: Sm motif of small nuclear ribonucleoproteins, SNRNP84 d1d3bb_
not modelled
5.6
8
Fold: Sm-like foldSuperfamily: Sm-like ribonucleoproteinsFamily: Sm motif of small nuclear ribonucleoproteins, SNRNP85 c3d6xA_
not modelled
5.5
27
PDB header: lyaseChain: A: PDB Molecule: (3r)-hydroxymyristoyl-[acyl-carrier-protein] dehydratase;PDBTitle: crystal structure of campylobacter jejuni fabz
86 d1p9ka_
not modelled
5.5
24
Fold: Alpha-L RNA-binding motifSuperfamily: Alpha-L RNA-binding motifFamily: YbcJ-like87 c3dzoA_
not modelled
5.3
16
PDB header: transferaseChain: A: PDB Molecule: rhoptry kinase domain;PDBTitle: crystal structure of a rhoptry kinase from toxoplasma gondii
88 d1yq2a3
not modelled
5.3
21
Fold: Galactose-binding domain-likeSuperfamily: Galactose-binding domain-likeFamily: beta-Galactosidase/glucuronidase, N-terminal domain89 c3cnrA_
not modelled
5.2
23
PDB header: unknown functionChain: A: PDB Molecule: type iv fimbriae assembly protein;PDBTitle: crystal structure of pilz (xac1133) from xanthomonas2 axonopodis pv citri
90 c1zyiA_
not modelled
5.2
11
PDB header: translationChain: A: PDB Molecule: methylosome subunit picln;PDBTitle: solution structure of icln, a multifunctional protein2 involved in regulatory mechanisms as different as cell3 volume regulation and rna splicing
91 d2cnda1
not modelled
5.2
33
Fold: Reductase/isomerase/elongation factor common domainSuperfamily: Riboflavin synthase domain-likeFamily: Ferredoxin reductase FAD-binding domain-like92 d2bmwa1
not modelled
5.0
33
Fold: Reductase/isomerase/elongation factor common domainSuperfamily: Riboflavin synthase domain-likeFamily: Ferredoxin reductase FAD-binding domain-like