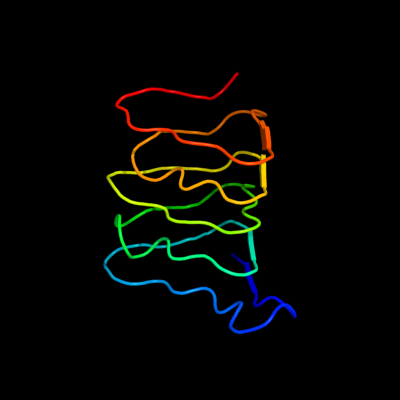
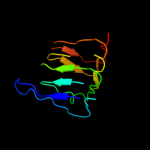
1 d2f9ca1
68.3
17
Fold: Single-stranded left-handed beta-helixSuperfamily: Trimeric LpxA-like enzymesFamily: YdcK-like2 c1fwyA_
54.2
19
PDB header: transferaseChain: A: PDB Molecule: udp-n-acetylglucosamine pyrophosphorylase;PDBTitle: crystal structure of n-acetylglucosamine 1-phosphate2 uridyltransferase bound to udp-glcnac
3 c1hf2A_
46.1
33
PDB header: cell division proteinChain: A: PDB Molecule: septum site-determining protein minc;PDBTitle: crystal structure of the bacterial cell-division inhibitor2 minc from t. maritima
4 c2i5kB_
43.3
13
PDB header: transferaseChain: B: PDB Molecule: utp--glucose-1-phosphate uridylyltransferase;PDBTitle: crystal structure of ugp1p
5 d1hf2a1
41.7
33
Fold: Single-stranded right-handed beta-helixSuperfamily: Cell-division inhibitor MinC, C-terminal domainFamily: Cell-division inhibitor MinC, C-terminal domain6 c3mc4A_
36.2
14
PDB header: transferaseChain: A: PDB Molecule: ww/rsp5/wwp domain:bacterial transferasePDBTitle: crystal structure of ww/rsp5/wwp domain: bacterial2 transferase hexapeptide repeat: serine o-acetyltransferase3 from brucella melitensis
7 c1hm8A_
32.6
15
PDB header: transferaseChain: A: PDB Molecule: udp-n-acetylglucosamine-1-phosphate uridyltransferase;PDBTitle: crystal structure of s.pneumoniae n-acetylglucosamine-1-phosphate2 uridyltransferase, glmu, bound to acetyl coenzyme a
8 d1o12a1
30.5
24
Fold: Composite domain of metallo-dependent hydrolasesSuperfamily: Composite domain of metallo-dependent hydrolasesFamily: N-acetylglucosamine-6-phosphate deacetylase, NagA9 d2icya1
30.0
22
Fold: Single-stranded left-handed beta-helixSuperfamily: Trimeric LpxA-like enzymesFamily: GlmU C-terminal domain-like10 c2qkxA_
28.9
8
PDB header: transferaseChain: A: PDB Molecule: bifunctional protein glmu;PDBTitle: n-acetyl glucosamine 1-phosphate uridyltransferase from mycobacterium2 tuberculosis complex with n-acetyl glucosamine 1-phosphate
11 c3b8eC_
28.7
15
PDB header: hydrolase/transport proteinChain: C: PDB Molecule: sodium/potassium-transporting atpase subunitPDBTitle: crystal structure of the sodium-potassium pump
12 c2i5nH_
28.6
15
PDB header: photosynthesisChain: H: PDB Molecule: reaction center protein h chain;PDBTitle: 1.96 a x-ray structure of photosynthetic reaction center from2 rhodopseudomonas viridis:crystals grown by microfluidic technique
13 c3brkX_
27.8
16
PDB header: transferaseChain: X: PDB Molecule: glucose-1-phosphate adenylyltransferase;PDBTitle: crystal structure of adp-glucose pyrophosphorylase from2 agrobacterium tumefaciens
14 c3d98A_
26.5
9
PDB header: transferaseChain: A: PDB Molecule: bifunctional protein glmu;PDBTitle: crystal structure of glmu from mycobacterium tuberculosis, ligand-free2 form
15 c2zxeA_
25.3
13
PDB header: hydrolase/transport proteinChain: A: PDB Molecule: na, k-atpase alpha subunit;PDBTitle: crystal structure of the sodium - potassium pump in the e2.2k+.pi2 state
16 c3f1xA_
20.4
19
PDB header: transferaseChain: A: PDB Molecule: serine acetyltransferase;PDBTitle: three dimensional structure of the serine acetyltransferase from2 bacteroides vulgatus, northeast structural genomics consortium target3 bvr62.
17 d1jk9b1
20.0
27
Fold: Immunoglobulin-like beta-sandwichSuperfamily: Cu,Zn superoxide dismutase-likeFamily: Cu,Zn superoxide dismutase-like18 c2z8gB_
18.9
14
PDB header: hydrolaseChain: B: PDB Molecule: isopullulanase;PDBTitle: aspergillus niger atcc9642 isopullulanase complexed with isopanose
19 d1ogmx2
16.9
18
Fold: Single-stranded right-handed beta-helixSuperfamily: Pectin lyase-likeFamily: Dextranase, catalytic domain20 c3mk7F_
15.6
25
PDB header: oxidoreductaseChain: F: PDB Molecule: cytochrome c oxidase, cbb3-type, subunit p;PDBTitle: the structure of cbb3 cytochrome oxidase
21 c3jx8B_
not modelled
15.1
20
PDB header: cell adhesionChain: B: PDB Molecule: putative lipoprotein;PDBTitle: crystal structure of putative lipid binding protein (yp_001304415.1)2 from parabacteroides distasonis atcc 8503 at 2.16 a resolution
22 c1t5eB_
not modelled
14.8
22
PDB header: transport proteinChain: B: PDB Molecule: multidrug resistance protein mexa;PDBTitle: the structure of mexa
23 c3gueB_
not modelled
14.2
16
PDB header: transferaseChain: B: PDB Molecule: utp-glucose-1-phosphate uridylyltransferase 2;PDBTitle: crystal structure of udp-glucose phosphorylase from trypanosoma2 brucei, (tb10.389.0330)
24 d1eysh2
not modelled
14.1
26
Fold: Single transmembrane helixSuperfamily: Photosystem II reaction centre subunit H, transmembrane regionFamily: Photosystem II reaction centre subunit H, transmembrane region25 c2q4jB_
not modelled
14.0
22
PDB header: transferaseChain: B: PDB Molecule: probable utp-glucose-1-phosphate uridylyltransferase 2;PDBTitle: ensemble refinement of the protein crystal structure of gene product2 from arabidopsis thaliana at3g03250, a putative udp-glucose3 pyrophosphorylase
26 c1k6nH_
not modelled
12.1
32
PDB header: photosynthesisChain: H: PDB Molecule: photosynthetic reaction center h subunit;PDBTitle: e(l212)a,d(l213)a double mutant structure of photosynthetic reaction2 center from rhodobacter sphaeroides
27 c1ogoX_
not modelled
11.3
17
PDB header: hydrolaseChain: X: PDB Molecule: dextranase;PDBTitle: dex49a from penicillium minioluteum complex with isomaltose
28 c3kdpD_
not modelled
9.1
12
PDB header: hydrolaseChain: D: PDB Molecule: sodium/potassium-transporting atpase subunit beta-1;PDBTitle: crystal structure of the sodium-potassium pump
29 c2v0hA_
not modelled
8.8
11
PDB header: transferaseChain: A: PDB Molecule: bifunctional protein glmu;PDBTitle: characterization of substrate binding and catalysis of the2 potential antibacterial target n-acetylglucosamine-1-3 phosphate uridyltransferase (glmu)
30 c1eysH_
not modelled
8.8
26
PDB header: electron transportChain: H: PDB Molecule: photosynthetic reaction center;PDBTitle: crystal structure of photosynthetic reaction center from a2 thermophilic bacterium, thermochromatium tepidum
31 c3ixzA_
not modelled
8.3
17
PDB header: hydrolaseChain: A: PDB Molecule: potassium-transporting atpase alpha;PDBTitle: pig gastric h+/k+-atpase complexed with aluminium fluoride
32 c2f1mA_
not modelled
8.2
20
PDB header: transport proteinChain: A: PDB Molecule: acriflavine resistance protein a;PDBTitle: conformational flexibility in the multidrug efflux system protein acra
33 d1ua7a1
not modelled
7.2
14
Fold: Glycosyl hydrolase domainSuperfamily: Glycosyl hydrolase domainFamily: alpha-Amylases, C-terminal beta-sheet domain34 c2aklA_
not modelled
6.6
17
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: phna-like protein pa0128;PDBTitle: solution structure for phn-a like protein pa0128 from2 pseudomonas aeruginosa
35 d2axtm1
not modelled
6.1
32
Fold: Single transmembrane helixSuperfamily: Photosystem II reaction center protein M, PsbMFamily: PsbM-like36 d2vnud3
not modelled
5.7
21
Fold: OB-foldSuperfamily: Nucleic acid-binding proteinsFamily: Cold shock DNA-binding domain-like37 d2akka1
not modelled
5.7
19
Fold: SH3-like barrelSuperfamily: Prokaryotic SH3-related domainFamily: PhnA-like38 d1srda_
not modelled
5.5
9
Fold: Immunoglobulin-like beta-sandwichSuperfamily: Cu,Zn superoxide dismutase-likeFamily: Cu,Zn superoxide dismutase-like