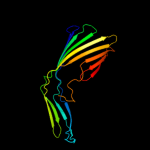
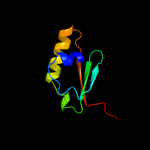
1 c2k0lA_
95.5
21
PDB header: membrane proteinChain: A: PDB Molecule: outer membrane protein a;PDBTitle: nmr structure of the transmembrane domain of the outer2 membrane protein a from klebsiella pneumoniae in dhpc3 micelles.
2 d1p4ta_
88.3
18
Fold: Transmembrane beta-barrelsSuperfamily: OMPA-likeFamily: Outer membrane protein3 d1g90a_
85.6
19
Fold: Transmembrane beta-barrelsSuperfamily: OMPA-likeFamily: Outer membrane protein4 c3bryB_
79.6
17
PDB header: transport proteinChain: B: PDB Molecule: tbux;PDBTitle: crystal structure of the ralstonia pickettii toluene2 transporter tbux
5 d1phoa_
75.0
14
Fold: Transmembrane beta-barrelsSuperfamily: PorinsFamily: Porin6 c3nb3C_
62.8
26
PDB header: virusChain: C: PDB Molecule: outer membrane protein a;PDBTitle: the host outer membrane proteins ompa and ompc are packed at specific2 sites in the shigella phage sf6 virion as structural components
7 c3dwoX_
60.9
12
PDB header: membrane proteinChain: X: PDB Molecule: probable outer membrane protein;PDBTitle: crystal structure of a pseudomonas aeruginosa fadl homologue
8 c3qraA_
55.2
24
PDB header: cell invasionChain: A: PDB Molecule: attachment invasion locus protein;PDBTitle: the crystal structure of ail, the attachment invasion locus protein of2 yersinia pestis
9 c2jmmA_
52.0
16
PDB header: membrane proteinChain: A: PDB Molecule: outer membrane protein a;PDBTitle: nmr solution structure of a minimal transmembrane beta-2 barrel platform protein
10 d1t16a_
50.4
13
Fold: Transmembrane beta-barrelsSuperfamily: PorinsFamily: Outer membrane protein transport protein11 d1qjpa_
47.7
22
Fold: Transmembrane beta-barrelsSuperfamily: OMPA-likeFamily: Outer membrane protein12 c2f1tB_
46.3
19
PDB header: membrane proteinChain: B: PDB Molecule: outer membrane protein w;PDBTitle: outer membrane protein ompw
13 d2zfga1
40.5
16
Fold: Transmembrane beta-barrelsSuperfamily: PorinsFamily: Porin14 c2x27X_
37.5
14
PDB header: membrane proteinChain: X: PDB Molecule: outer membrane protein oprg;PDBTitle: crystal structure of the outer membrane protein oprg from2 pseudomonas aeruginosa
15 d1ynja1
37.3
18
Fold: DCoH-likeSuperfamily: RBP11-like subunits of RNA polymeraseFamily: RNA polymerase alpha subunit dimerisation domain16 c2jvfA_
36.4
12
PDB header: de novo proteinChain: A: PDB Molecule: de novo protein m7;PDBTitle: solution structure of m7, a computationally-designed2 artificial protein
17 c2lhfA_
33.6
20
PDB header: membrane proteinChain: A: PDB Molecule: outer membrane protein h1;PDBTitle: solution structure of outer membrane protein h (oprh) from p.2 aeruginosa in dhpc micelles
18 c1qysA_
29.3
22
PDB header: de novo proteinChain: A: PDB Molecule: top7;PDBTitle: crystal structure of top7: a computationally designed2 protein with a novel fold
19 c3brzA_
28.0
13
PDB header: transport proteinChain: A: PDB Molecule: todx;PDBTitle: crystal structure of the pseudomonas putida toluene2 transporter todx
20 c2e0gA_
25.9
11
PDB header: replicationChain: A: PDB Molecule: chromosomal replication initiator protein dnaa;PDBTitle: dnaa n-terminal domain
21 c3a2rX_
not modelled
25.5
14
PDB header: membrane proteinChain: X: PDB Molecule: outer membrane protein ii;PDBTitle: crystal structure of outer membrane protein porb from neisseria2 meningitidis
22 d2fgqx1
not modelled
21.5
16
Fold: Transmembrane beta-barrelsSuperfamily: PorinsFamily: Porin23 c2i9zB_
not modelled
20.2
22
PDB header: structural genomics, unknown functionChain: B: PDB Molecule: putative septation protein spovg;PDBTitle: structural genomics, the crystal structure of full-length spovg from2 staphylococcus epidermidis atcc 12228
24 d2i9xa1
not modelled
20.2
22
Fold: SpoVG-likeSuperfamily: SpoVG-likeFamily: SpoVG-like25 d1uynx_
not modelled
15.9
11
Fold: Transmembrane beta-barrelsSuperfamily: AutotransporterFamily: Autotransporter26 d1bdfa1
not modelled
14.0
33
Fold: DCoH-likeSuperfamily: RBP11-like subunits of RNA polymeraseFamily: RNA polymerase alpha subunit dimerisation domain27 d1umha_
not modelled
10.3
25
Fold: Galactose-binding domain-likeSuperfamily: Galactose-binding domain-likeFamily: F-box associated region, FBA28 d1u69a_
not modelled
10.3
23
Fold: Glyoxalase/Bleomycin resistance protein/Dihydroxybiphenyl dioxygenaseSuperfamily: Glyoxalase/Bleomycin resistance protein/Dihydroxybiphenyl dioxygenaseFamily: 3-demethylubiquinone-9 3-methyltransferase29 d1kpta_
not modelled
9.5
35
Fold: Yeast killer toxinsSuperfamily: Yeast killer toxinsFamily: Virally encoded KP4 toxin30 c2gjhA_
not modelled
9.1
30
PDB header: de novo proteinChain: A: PDB Molecule: designed protein;PDBTitle: nmr structure of cfr (c-terminal fragment of2 computationally designed novel-topology protein top7)
31 d2gufa1
not modelled
9.0
18
Fold: Transmembrane beta-barrelsSuperfamily: PorinsFamily: Ligand-gated protein channel32 c2x4mD_
not modelled
8.6
19
PDB header: hydrolaseChain: D: PDB Molecule: coagulase/fibrinolysin;PDBTitle: yersinia pestis plasminogen activator pla
33 c3qwnC_
not modelled
8.5
23
PDB header: antitoxinChain: C: PDB Molecule: hypothetical nigd-like protein;PDBTitle: crystal structure of a hypothetical nigd-like protein (baccac_03262)2 from bacteroides caccae at 2.42 a resolution
34 d1tdha3
not modelled
8.0
20
Fold: Glucocorticoid receptor-like (DNA-binding domain)Superfamily: Glucocorticoid receptor-like (DNA-binding domain)Family: C-terminal, Zn-finger domain of MutM-like DNA repair proteins35 c2djmA_
not modelled
7.7
16
PDB header: sugar binding proteinChain: A: PDB Molecule: glucoamylase a;PDBTitle: solution structure of n-terminal starch-binding domain of2 glucoamylase from rhizopus oryzae
36 c3qv0A_
not modelled
7.7
16
PDB header: protein bindingChain: A: PDB Molecule: mitochondrial acidic protein mam33;PDBTitle: crystal structure of saccharomyces cerevisiae mam33
37 d1smya1
not modelled
7.7
20
Fold: DCoH-likeSuperfamily: RBP11-like subunits of RNA polymeraseFamily: RNA polymerase alpha subunit dimerisation domain38 d1f2va_
not modelled
7.6
18
Fold: Flavodoxin-likeSuperfamily: Precorrin-8X methylmutase CbiC/CobHFamily: Precorrin-8X methylmutase CbiC/CobH39 d1txka2
not modelled
7.0
18
Fold: SupersandwichSuperfamily: Galactose mutarotase-likeFamily: MdoG-like40 d1kkea2
not modelled
7.0
20
Fold: Virus attachment protein globular domainSuperfamily: Virus attachment protein globular domainFamily: Reovirus attachment protein sigma 1 head domain41 d1smyc_
not modelled
6.9
32
Fold: beta and beta-prime subunits of DNA dependent RNA-polymeraseSuperfamily: beta and beta-prime subunits of DNA dependent RNA-polymeraseFamily: RNA-polymerase beta42 c3e7dC_
not modelled
6.9
18
PDB header: isomeraseChain: C: PDB Molecule: cobh, precorrin-8x methylmutase;PDBTitle: crystal structure of precorrin-8x methyl mutase cbic/cobh from2 brucella melitensis
43 c2r28D_
not modelled
6.9
18
PDB header: metal binding protein/hydrolaseChain: D: PDB Molecule: serine/threonine-protein phosphatase 2bPDBTitle: the complex structure of calmodulin bound to a calcineurin2 peptide
44 c1txkA_
not modelled
6.7
18
PDB header: biosynthetic proteinChain: A: PDB Molecule: glucans biosynthesis protein g;PDBTitle: crystal structure of escherichia coli opgg
45 c2jziB_
not modelled
6.7
20
PDB header: metal binding proteinChain: B: PDB Molecule: serine/threonine-protein phosphatase 2bPDBTitle: structure of calmodulin complexed with the calmodulin2 binding domain of calcineurin
46 c3cjtP_
not modelled
6.5
23
PDB header: transferase/ribosomal proteinChain: P: PDB Molecule: 50s ribosomal protein l11;PDBTitle: ribosomal protein l11 methyltransferase (prma) in complex with2 dimethylated ribosomal protein l11
47 d1kk1a2
not modelled
6.5
25
Fold: Elongation factor/aminomethyltransferase common domainSuperfamily: EF-Tu/eEF-1alpha/eIF2-gamma C-terminal domainFamily: EF-Tu/eEF-1alpha/eIF2-gamma C-terminal domain48 c2rqeA_
not modelled
6.4
75
PDB header: sugar binding proteinChain: A: PDB Molecule: beta-1,3-glucan-binding protein;PDBTitle: solution structure of the silkworm bgrp/gnbp3 n-terminal2 domain reveals the mechanism for b-1,3-glucan specific3 recognition
49 c3fhhA_
not modelled
6.4
6
PDB header: membrane proteinChain: A: PDB Molecule: outer membrane heme receptor shua;PDBTitle: crystal structure of the heme/hemoglobin outer membrane2 transporter shua from shigella dysenteriae
50 c2afvB_
not modelled
6.3
24
PDB header: isomeraseChain: B: PDB Molecule: cobalamin biosynthesis precorrin isomerase;PDBTitle: the crystal structure of putative precorrin isomerase cbic2 in cobalamin biosynthesis
51 c1uv7A_
not modelled
6.2
10
PDB header: transportChain: A: PDB Molecule: general secretion pathway protein m;PDBTitle: periplasmic domain of epsm from vibrio cholerae
52 d1uv7a_
not modelled
6.2
10
Fold: RRF/tRNA synthetase additional domain-likeSuperfamily: General secretion pathway protein M, EpsMFamily: General secretion pathway protein M, EpsM53 d2cura2
not modelled
6.2
55
Fold: Glucocorticoid receptor-like (DNA-binding domain)Superfamily: Glucocorticoid receptor-like (DNA-binding domain)Family: LIM domain54 c3ie4A_
not modelled
6.2
63
PDB header: immune systemChain: A: PDB Molecule: gram-negative binding protein 3;PDBTitle: b-glucan binding domain of drosophila gnbp3 defines a novel2 family of pattern recognition receptor
55 d2jnaa1
not modelled
6.0
24
Fold: Dodecin subunit-likeSuperfamily: YdgH-likeFamily: YdgH-like56 c3iydC_
not modelled
5.7
27
PDB header: transcription/dnaChain: C: PDB Molecule: dna-directed rna polymerase subunit beta;PDBTitle: three-dimensional em structure of an intact activator-dependent2 transcription initiation complex
57 c2khaA_
not modelled
5.5
44
PDB header: sugar binding proteinChain: A: PDB Molecule: beta-1,3-glucan-binding protein;PDBTitle: solution structure of a pathogen recognition domain from a2 lepidopteran insect, plodia interpunctella
58 c1kkeA_
not modelled
5.4
20
PDB header: viral proteinChain: A: PDB Molecule: sigma 1 protein;PDBTitle: crystal structure of reovirus attachment protein sigma12 trimer
59 d2d8za1
not modelled
5.4
55
Fold: Glucocorticoid receptor-like (DNA-binding domain)Superfamily: Glucocorticoid receptor-like (DNA-binding domain)Family: LIM domain60 d2cnaa_
not modelled
5.4
19
Fold: Concanavalin A-like lectins/glucanasesSuperfamily: Concanavalin A-like lectins/glucanasesFamily: Legume lectins61 d2gr8a1
not modelled
5.4
18
Fold: Pili subunitsSuperfamily: Pili subunitsFamily: YadA C-terminal domain-like62 c3jsrA_
not modelled
5.4
33
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: all0216 protein;PDBTitle: x-ray structure of all0216 protein from nostoc sp. pcc 7120 at the2 resolution 1.8a. northeast structural genomics consortium target3 nsr236
63 d2pwwa1
not modelled
5.3
21
Fold: TBP-likeSuperfamily: YugN-likeFamily: YugN-like64 c3e8vA_
not modelled
5.2
35
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: possible transglutaminase-family protein;PDBTitle: crystal structure of a possible transglutaminase-family2 protein proteolytic fragment from bacteroides fragilis
65 d1ou0a_
not modelled
5.2
35
Fold: Flavodoxin-likeSuperfamily: Precorrin-8X methylmutase CbiC/CobHFamily: Precorrin-8X methylmutase CbiC/CobH66 c3nsgA_
not modelled
5.2
15
PDB header: membrane proteinChain: A: PDB Molecule: outer membrane protein f;PDBTitle: crystal structure of ompf, an outer membrane protein from salmonella2 typhi
67 c3o27B_
not modelled
5.2
17
PDB header: dna binding proteinChain: B: PDB Molecule: putative uncharacterized protein;PDBTitle: the crystal structure of c68 from the hybrid virus-plasmid pssvx
68 c3qq2C_
not modelled
5.2
15
PDB header: membrane protein/protein transportChain: C: PDB Molecule: brka autotransporter;PDBTitle: crystal structure of the beta domain of the bordetella autotransporter2 brka
69 d3prna_
not modelled
5.1
11
Fold: Transmembrane beta-barrelsSuperfamily: PorinsFamily: Porin70 d3cjsb1
not modelled
5.1
27
Fold: Ribosomal L11/L12e N-terminal domainSuperfamily: Ribosomal L11/L12e N-terminal domainFamily: Ribosomal L11/L12e N-terminal domain71 d1h9wa_
not modelled
5.0
19
Fold: Concanavalin A-like lectins/glucanasesSuperfamily: Concanavalin A-like lectins/glucanasesFamily: Legume lectins72 d2okga1
not modelled
5.0
13
Fold: NagB/RpiA/CoA transferase-likeSuperfamily: NagB/RpiA/CoA transferase-likeFamily: SorC sugar-binding domain-like