| 1 | c3fd0B_
|
|
 |
100.0 |
15 |
PDB header:lyase
Chain: B: PDB Molecule:putative cystathionine beta-lyase involved in aluminum
PDBTitle: crystal structure of putative cystathionine beta-lyase involved in2 aluminum resistance (np_470671.1) from listeria innocua at 2.12 a3 resolution
|
| 2 | c3ht4B_
|
|
 |
100.0 |
20 |
PDB header:lyase
Chain: B: PDB Molecule:aluminum resistance protein;
PDBTitle: crystal structure of the q81a77_baccr protein from bacillus2 cereus. northeast structural genomics consortium target3 bcr213
|
| 3 | c2cb1A_
|
|
 |
100.0 |
21 |
PDB header:lyase
Chain: A: PDB Molecule:o-acetyl homoserine sulfhydrylase;
PDBTitle: crystal structure of o-actetyl homoserine sulfhydrylase2 from thermus thermophilus hb8,oah2.
|
| 4 | c3ndnC_
|
|
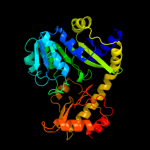 |
100.0 |
20 |
PDB header:lyase
Chain: C: PDB Molecule:o-succinylhomoserine sulfhydrylase;
PDBTitle: crystal structure of o-succinylhomoserine sulfhydrylase from2 mycobacterium tuberculosis covalently bound to pyridoxal-5-phosphate
|
| 5 | c3hvyC_
|
|
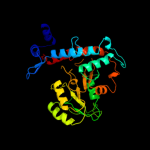 |
100.0 |
17 |
PDB header:lyase
Chain: C: PDB Molecule:cystathionine beta-lyase family protein, ynbb b.subtilis
PDBTitle: crystal structure of putative cystathionine beta-lyase involved in2 aluminum resistance (np_348457.1) from clostridium acetobutylicum at3 2.00 a resolution
|
| 6 | d1qgna_
|
|
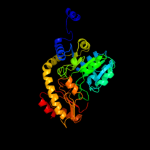 |
100.0 |
20 |
Fold:PLP-dependent transferase-like
Superfamily:PLP-dependent transferases
Family:Cystathionine synthase-like |
| 7 | c1ibjC_
|
|
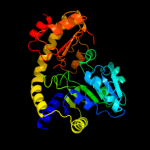 |
100.0 |
18 |
PDB header:lyase
Chain: C: PDB Molecule:cystathionine beta-lyase;
PDBTitle: crystal structure of cystathionine beta-lyase from arabidopsis2 thaliana
|
| 8 | d1ibja_
|
|
 |
100.0 |
18 |
Fold:PLP-dependent transferase-like
Superfamily:PLP-dependent transferases
Family:Cystathionine synthase-like |
| 9 | c3gwpA_
|
|
 |
100.0 |
17 |
PDB header:lyase
Chain: A: PDB Molecule:carbon-sulfur lyase involved in aluminum resistance;
PDBTitle: crystal structure of carbon-sulfur lyase involved in aluminum2 resistance (yp_878183.1) from clostridium novyi nt at 2.90 a3 resolution
|
| 10 | c2gqnB_
|
|
 |
100.0 |
17 |
PDB header:lyase
Chain: B: PDB Molecule:cystathionine beta-lyase;
PDBTitle: cystathionine beta-lyase (cbl) from escherichia coli in complex with2 n-hydrazinocarbonylmethyl-2-nitro-benzamide
|
| 11 | c3bcxA_
|
|
 |
100.0 |
15 |
PDB header:transferase
Chain: A: PDB Molecule:cdp-6-deoxy-l-threo-d-glycero-4-hexulose-3-
PDBTitle: e1 dehydrase
|
| 12 | d1y4ia1
|
|
 |
100.0 |
19 |
Fold:PLP-dependent transferase-like
Superfamily:PLP-dependent transferases
Family:Cystathionine synthase-like |
| 13 | d1cl1a_
|
|
 |
100.0 |
18 |
Fold:PLP-dependent transferase-like
Superfamily:PLP-dependent transferases
Family:Cystathionine synthase-like |
| 14 | d2ctza1
|
|
 |
100.0 |
17 |
Fold:PLP-dependent transferase-like
Superfamily:PLP-dependent transferases
Family:Cystathionine synthase-like |
| 15 | c2c7tA_
|
|
 |
100.0 |
17 |
PDB header:transferase
Chain: A: PDB Molecule:glutamine-2-deoxy-scyllo-inosose
PDBTitle: crystal structure of the plp-bound form of btrr,2 a dual functional aminotransferase involved in butirosin3 biosynthesis.
|
| 16 | c3aemD_
|
|
 |
100.0 |
14 |
PDB header:lyase
Chain: D: PDB Molecule:methionine gamma-lyase;
PDBTitle: reaction intermediate structure of entamoeba histolytica methionine2 gamma-lyase 1 containing michaelis complex and methionine imine-3 pyridoxamine-5'-phosphate
|
| 17 | c2nmpC_
|
|
 |
100.0 |
18 |
PDB header:lyase
Chain: C: PDB Molecule:cystathionine gamma-lyase;
PDBTitle: crystal structure of human cystathionine gamma lyase
|
| 18 | c1i41J_
|
|
 |
100.0 |
20 |
PDB header:lyase
Chain: J: PDB Molecule:cystathionine gamma-synthase;
PDBTitle: cystathionine gamma-synthase in complex with the inhibitor2 appa
|
| 19 | c3e6gA_
|
|
 |
100.0 |
20 |
PDB header:lyase
Chain: A: PDB Molecule:cystathionine gamma-lyase-like protein;
PDBTitle: crystal structure of xometc, a cystathionine c-lyase-like2 protein from xanthomonas oryzae pv.oryzae
|
| 20 | d1n8pa_
|
|
 |
100.0 |
19 |
Fold:PLP-dependent transferase-like
Superfamily:PLP-dependent transferases
Family:Cystathionine synthase-like |
| 21 | d1e5ea_ |
|
not modelled |
100.0 |
15 |
Fold:PLP-dependent transferase-like
Superfamily:PLP-dependent transferases
Family:Cystathionine synthase-like |
| 22 | d1cs1a_ |
|
not modelled |
100.0 |
19 |
Fold:PLP-dependent transferase-like
Superfamily:PLP-dependent transferases
Family:Cystathionine synthase-like |
| 23 | c2r0tA_ |
|
not modelled |
100.0 |
15 |
PDB header:transferase
Chain: A: PDB Molecule:pyridoxamine 5-phosphate-dependent dehydrase;
PDBTitle: crystal sructure of gdp-4-keto-6-deoxymannose-3-dehydratase2 with a trapped plp-glutamate geminal diamine
|
| 24 | d1wyub1 |
|
not modelled |
100.0 |
18 |
Fold:PLP-dependent transferase-like
Superfamily:PLP-dependent transferases
Family:Glycine dehydrogenase subunits (GDC-P) |
| 25 | c3qi6B_ |
|
not modelled |
100.0 |
19 |
PDB header:lyase
Chain: B: PDB Molecule:cystathionine gamma-synthase metb (cgs);
PDBTitle: crystal structure of cystathionine gamma-synthase metb (cgs) from2 mycobacterium ulcerans agy99
|
| 26 | d1gc0a_ |
|
not modelled |
100.0 |
21 |
Fold:PLP-dependent transferase-like
Superfamily:PLP-dependent transferases
Family:Cystathionine synthase-like |
| 27 | d1mdoa_ |
|
not modelled |
100.0 |
17 |
Fold:PLP-dependent transferase-like
Superfamily:PLP-dependent transferases
Family:GABA-aminotransferase-like |
| 28 | c2po3B_ |
|
not modelled |
100.0 |
16 |
PDB header:transferase
Chain: B: PDB Molecule:4-dehydrase;
PDBTitle: crystal structure analysis of desi in the presence of its2 tdp-sugar product
|
| 29 | d1b9ha_ |
|
not modelled |
100.0 |
17 |
Fold:PLP-dependent transferase-like
Superfamily:PLP-dependent transferases
Family:GABA-aminotransferase-like |
| 30 | d1wyua1 |
|
not modelled |
100.0 |
15 |
Fold:PLP-dependent transferase-like
Superfamily:PLP-dependent transferases
Family:Glycine dehydrogenase subunits (GDC-P) |
| 31 | c3ju7B_ |
|
not modelled |
100.0 |
13 |
PDB header:transferase
Chain: B: PDB Molecule:putative plp-dependent aminotransferase;
PDBTitle: crystal structure of putative plp-dependent aminotransferase2 (np_978343.1) from bacillus cereus atcc 10987 at 2.19 a resolution
|
| 32 | c3pj0D_ |
|
not modelled |
100.0 |
13 |
PDB header:lyase
Chain: D: PDB Molecule:lmo0305 protein;
PDBTitle: crystal structure of a putative l-allo-threonine aldolase (lmo0305)2 from listeria monocytogenes egd-e at 1.80 a resolution
|
| 33 | d2fnua1 |
|
not modelled |
100.0 |
14 |
Fold:PLP-dependent transferase-like
Superfamily:PLP-dependent transferases
Family:GABA-aminotransferase-like |
| 34 | d1o69a_ |
|
not modelled |
100.0 |
13 |
Fold:PLP-dependent transferase-like
Superfamily:PLP-dependent transferases
Family:GABA-aminotransferase-like |
| 35 | c2ogeC_ |
|
not modelled |
99.9 |
16 |
PDB header:transferase
Chain: C: PDB Molecule:transaminase;
PDBTitle: x-ray structure of s. venezuelae desv in its internal2 aldimine form
|
| 36 | c3nx3A_ |
|
not modelled |
99.9 |
10 |
PDB header:transferase
Chain: A: PDB Molecule:acetylornithine aminotransferase;
PDBTitle: crystal structure of acetylornithine aminotransferase (argd) from2 campylobacter jejuni
|
| 37 | c3mafB_ |
|
not modelled |
99.9 |
16 |
PDB header:lyase
Chain: B: PDB Molecule:sphingosine-1-phosphate lyase;
PDBTitle: crystal structure of stspl (asymmetric form)
|
| 38 | c2w8wA_ |
|
not modelled |
99.9 |
15 |
PDB header:transferase
Chain: A: PDB Molecule:serine palmitoyltransferase;
PDBTitle: n100y spt with plp-ser
|
| 39 | c3dr4B_ |
|
not modelled |
99.9 |
14 |
PDB header:transferase
Chain: B: PDB Molecule:putative perosamine synthetase;
PDBTitle: gdp-perosamine synthase k186a mutant from caulobacter2 crescentus with bound sugar ligand
|
| 40 | c3nysA_ |
|
not modelled |
99.9 |
15 |
PDB header:transferase
Chain: A: PDB Molecule:aminotransferase wbpe;
PDBTitle: x-ray structure of the k185a mutant of wbpe (wlbe) from pseudomonas2 aeruginosa in complex with plp at 1.45 angstrom resolution
|
| 41 | d2byla1 |
|
not modelled |
99.9 |
9 |
Fold:PLP-dependent transferase-like
Superfamily:PLP-dependent transferases
Family:GABA-aminotransferase-like |
| 42 | c3hbxB_ |
|
not modelled |
99.9 |
15 |
PDB header:lyase
Chain: B: PDB Molecule:glutamate decarboxylase 1;
PDBTitle: crystal structure of gad1 from arabidopsis thaliana
|
| 43 | c3ruyB_ |
|
not modelled |
99.9 |
12 |
PDB header:transferase
Chain: B: PDB Molecule:ornithine aminotransferase;
PDBTitle: crystal structure of the ornithine-oxo acid transaminase rocd from2 bacillus anthracis
|
| 44 | c1oatB_ |
|
not modelled |
99.9 |
10 |
PDB header:aminotransferase
Chain: B: PDB Molecule:ornithine aminotransferase;
PDBTitle: ornithine aminotransferase
|
| 45 | c3caiA_ |
|
not modelled |
99.9 |
15 |
PDB header:transferase
Chain: A: PDB Molecule:possible aminotransferase;
PDBTitle: crystal structure of mycobacterium tuberculosis rv3778c2 protein
|
| 46 | d1z7da1 |
|
not modelled |
99.9 |
11 |
Fold:PLP-dependent transferase-like
Superfamily:PLP-dependent transferases
Family:GABA-aminotransferase-like |
| 47 | c3mc6C_ |
|
not modelled |
99.9 |
12 |
PDB header:lyase
Chain: C: PDB Molecule:sphingosine-1-phosphate lyase;
PDBTitle: crystal structure of scdpl1
|
| 48 | d1bs0a_ |
|
not modelled |
99.9 |
19 |
Fold:PLP-dependent transferase-like
Superfamily:PLP-dependent transferases
Family:GABA-aminotransferase-like |
| 49 | c3ri6A_ |
|
not modelled |
99.9 |
18 |
PDB header:transferase
Chain: A: PDB Molecule:o-acetylhomoserine sulfhydrylase;
PDBTitle: a novel mechanism of sulfur transfer catalyzed by o-acetylhomoserine2 sulfhydrylase in methionine biosynthetic pathway of wolinella3 succinogenes
|
| 50 | c3uwcA_ |
|
not modelled |
99.9 |
14 |
PDB header:transferase
Chain: A: PDB Molecule:nucleotide-sugar aminotransferase;
PDBTitle: structure of an aminotransferase (degt-dnrj-eryc1-strs family) from2 coxiella burnetii in complex with pmp
|
| 51 | c3l44A_ |
|
not modelled |
99.9 |
15 |
PDB header:isomerase
Chain: A: PDB Molecule:glutamate-1-semialdehyde 2,1-aminomutase 1;
PDBTitle: crystal structure of bacillus anthracis heml-1, glutamate semialdehyde2 aminotransferase
|
| 52 | d1jf9a_ |
|
not modelled |
99.9 |
16 |
Fold:PLP-dependent transferase-like
Superfamily:PLP-dependent transferases
Family:Cystathionine synthase-like |
| 53 | d1c4ka2 |
|
not modelled |
99.9 |
16 |
Fold:PLP-dependent transferase-like
Superfamily:PLP-dependent transferases
Family:Ornithine decarboxylase major domain |
| 54 | c3frkB_ |
|
not modelled |
99.9 |
13 |
PDB header:transferase
Chain: B: PDB Molecule:qdtb;
PDBTitle: x-ray structure of qdtb from t. thermosaccharolyticum in2 complex with a plp:tdp-3-aminoquinovose aldimine
|
| 55 | c3h7fB_ |
|
not modelled |
99.9 |
19 |
PDB header:transferase
Chain: B: PDB Molecule:serine hydroxymethyltransferase 1;
PDBTitle: crystal structure of serine hydroxymethyltransferase from2 mycobacterium tuberculosis
|
| 56 | d1vefa1 |
|
not modelled |
99.9 |
15 |
Fold:PLP-dependent transferase-like
Superfamily:PLP-dependent transferases
Family:GABA-aminotransferase-like |
| 57 | c3b46B_ |
|
not modelled |
99.9 |
14 |
PDB header:transferase
Chain: B: PDB Molecule:aminotransferase bna3;
PDBTitle: crystal structure of bna3p, a putative kynurenine2 aminotransferase from saccharomyces cerevisiae
|
| 58 | d2bwna1 |
|
not modelled |
99.9 |
12 |
Fold:PLP-dependent transferase-like
Superfamily:PLP-dependent transferases
Family:GABA-aminotransferase-like |
| 59 | c3e9kA_ |
|
not modelled |
99.9 |
15 |
PDB header:hydrolase
Chain: A: PDB Molecule:kynureninase;
PDBTitle: crystal structure of homo sapiens kynureninase-3-hydroxyhippuric acid2 inhibitor complex
|
| 60 | c3dodA_ |
|
not modelled |
99.9 |
12 |
PDB header:transferase
Chain: A: PDB Molecule:adenosylmethionine-8-amino-7-oxononanoate aminotransferase;
PDBTitle: crystal structure of plp bound 7,8-diaminopelargonic acid synthase in2 bacillus subtilis
|
| 61 | d1s0aa_ |
|
not modelled |
99.9 |
10 |
Fold:PLP-dependent transferase-like
Superfamily:PLP-dependent transferases
Family:GABA-aminotransferase-like |
| 62 | c3n0lA_ |
|
not modelled |
99.9 |
16 |
PDB header:transferase
Chain: A: PDB Molecule:serine hydroxymethyltransferase;
PDBTitle: crystal structure of serine hydroxymethyltransferase from2 campylobacter jejuni
|
| 63 | c2pb2B_ |
|
not modelled |
99.9 |
12 |
PDB header:transferase
Chain: B: PDB Molecule:acetylornithine/succinyldiaminopimelate aminotransferase;
PDBTitle: structure of biosynthetic n-acetylornithine aminotransferase from2 salmonella typhimurium: studies on substrate specificity and3 inhibitor binding
|
| 64 | d1pffa_ |
|
not modelled |
99.9 |
16 |
Fold:PLP-dependent transferase-like
Superfamily:PLP-dependent transferases
Family:Cystathionine synthase-like |
| 65 | c3a8uX_ |
|
not modelled |
99.9 |
13 |
PDB header:transferase
Chain: X: PDB Molecule:omega-amino acid--pyruvate aminotransferase;
PDBTitle: crystal structure of omega-amino acid:pyruvate aminotransferase
|
| 66 | d1fc4a_ |
|
not modelled |
99.9 |
15 |
Fold:PLP-dependent transferase-like
Superfamily:PLP-dependent transferases
Family:GABA-aminotransferase-like |
| 67 | c3tqxA_ |
|
not modelled |
99.9 |
14 |
PDB header:transferase
Chain: A: PDB Molecule:2-amino-3-ketobutyrate coenzyme a ligase;
PDBTitle: structure of the 2-amino-3-ketobutyrate coenzyme a ligase (kbl) from2 coxiella burnetii
|
| 68 | d1c7ga_ |
|
not modelled |
99.9 |
11 |
Fold:PLP-dependent transferase-like
Superfamily:PLP-dependent transferases
Family:Beta-eliminating lyases |
| 69 | c2ordA_ |
|
not modelled |
99.9 |
12 |
PDB header:transferase
Chain: A: PDB Molecule:acetylornithine aminotransferase;
PDBTitle: crystal structure of acetylornithine aminotransferase (ec 2.6.1.11)2 (acoat) (tm1785) from thermotoga maritima at 1.40 a resolution
|
| 70 | c3hmuA_ |
|
not modelled |
99.9 |
11 |
PDB header:transferase
Chain: A: PDB Molecule:aminotransferase, class iii;
PDBTitle: crystal structure of a class iii aminotransferase from2 silicibacter pomeroyi
|
| 71 | d2gsaa_ |
|
not modelled |
99.9 |
15 |
Fold:PLP-dependent transferase-like
Superfamily:PLP-dependent transferases
Family:GABA-aminotransferase-like |
| 72 | c3i4jC_ |
|
not modelled |
99.9 |
9 |
PDB header:transferase
Chain: C: PDB Molecule:aminotransferase, class iii;
PDBTitle: crystal structure of aminotransferase, class iii from2 deinococcus radiodurans
|
| 73 | c2eh6A_ |
|
not modelled |
99.9 |
11 |
PDB header:transferase
Chain: A: PDB Molecule:acetylornithine aminotransferase;
PDBTitle: crystal structure of acetylornithine aminotransferase from aquifex2 aeolicus vf5
|
| 74 | c3a2bA_ |
|
not modelled |
99.9 |
15 |
PDB header:transferase
Chain: A: PDB Molecule:serine palmitoyltransferase;
PDBTitle: crystal structure of serine palmitoyltransferase from sphingobacterium2 multivorum with substrate l-serine
|
| 75 | c1z7dE_ |
|
not modelled |
99.9 |
11 |
PDB header:transferase
Chain: E: PDB Molecule:ornithine aminotransferase;
PDBTitle: ornithine aminotransferase py00104 from plasmodium yoelii
|
| 76 | d1zoda1 |
|
not modelled |
99.9 |
13 |
Fold:PLP-dependent transferase-like
Superfamily:PLP-dependent transferases
Family:GABA-aminotransferase-like |
| 77 | d2aeua1 |
|
not modelled |
99.9 |
17 |
Fold:PLP-dependent transferase-like
Superfamily:PLP-dependent transferases
Family:SelA-like |
| 78 | d3bc8a1 |
|
not modelled |
99.9 |
13 |
Fold:PLP-dependent transferase-like
Superfamily:PLP-dependent transferases
Family:SepSecS-like |
| 79 | d1kl1a_ |
|
not modelled |
99.9 |
17 |
Fold:PLP-dependent transferase-like
Superfamily:PLP-dependent transferases
Family:GABA-aminotransferase-like |
| 80 | c3lwsF_ |
|
not modelled |
99.9 |
14 |
PDB header:lyase
Chain: F: PDB Molecule:aromatic amino acid beta-eliminating
PDBTitle: crystal structure of putative aromatic amino acid beta-2 eliminating lyase/threonine aldolase. (yp_001813866.1) from3 exiguobacterium sp. 255-15 at 2.00 a resolution
|
| 81 | c2x3lA_ |
|
not modelled |
99.9 |
14 |
PDB header:lyase
Chain: A: PDB Molecule:orn/lys/arg decarboxylase family protein;
PDBTitle: crystal structure of the orn_lys_arg decarboxylase family2 protein sar0482 from methicillin-resistant staphylococcus3 aureus
|
| 82 | c3e2yB_ |
|
not modelled |
99.9 |
11 |
PDB header:transferase, lyase
Chain: B: PDB Molecule:kynurenine-oxoglutarate transaminase 3;
PDBTitle: crystal structure of mouse kynurenine aminotransferase iii in complex2 with glutamine
|
| 83 | c3jtxB_ |
|
not modelled |
99.9 |
14 |
PDB header:transferase
Chain: B: PDB Molecule:aminotransferase;
PDBTitle: crystal structure of aminotransferase (np_283882.1) from neisseria2 meningitidis z2491 at 1.91 a resolution
|
| 84 | c3bs8A_ |
|
not modelled |
99.9 |
15 |
PDB header:isomerase
Chain: A: PDB Molecule:glutamate-1-semialdehyde 2,1-aminomutase;
PDBTitle: crystal structure of glutamate 1-semialdehyde aminotransferase2 complexed with pyridoxamine-5'-phosphate from bacillus subtilis
|
| 85 | c3nuiA_ |
|
not modelled |
99.9 |
13 |
PDB header:transferase
Chain: A: PDB Molecule:pyruvate transaminase;
PDBTitle: crystal structure of omega-transferase from vibrio fluvialis js17
|
| 86 | c2cy8A_ |
|
not modelled |
99.9 |
14 |
PDB header:transferase
Chain: A: PDB Molecule:d-phenylglycine aminotransferase;
PDBTitle: crystal structure of d-phenylglycine aminotransferase (d-phgat) from2 pseudomonas strutzeri st-201
|
| 87 | d1qz9a_ |
|
not modelled |
99.9 |
17 |
Fold:PLP-dependent transferase-like
Superfamily:PLP-dependent transferases
Family:Cystathionine synthase-like |
| 88 | c2dkjB_ |
|
not modelled |
99.9 |
15 |
PDB header:transferase
Chain: B: PDB Molecule:serine hydroxymethyltransferase;
PDBTitle: crystal structure of t.th.hb8 serine hydroxymethyltransferase
|
| 89 | d2r5ea1 |
|
not modelled |
99.9 |
16 |
Fold:PLP-dependent transferase-like
Superfamily:PLP-dependent transferases
Family:AAT-like |
| 90 | c2e7uA_ |
|
not modelled |
99.9 |
18 |
PDB header:isomerase
Chain: A: PDB Molecule:glutamate-1-semialdehyde 2,1-aminomutase;
PDBTitle: crystal structure of glutamate-1-semialdehyde 2,1-aminomutase from2 thermus thermophilus hb8
|
| 91 | c2zsmA_ |
|
not modelled |
99.9 |
17 |
PDB header:isomerase
Chain: A: PDB Molecule:glutamate-1-semialdehyde 2,1-aminomutase;
PDBTitle: crystal structure of glutamate-1-semialdehyde 2,1-2 aminomutase from aeropyrum pernix, hexagonal form
|
| 92 | d1m6sa_ |
|
not modelled |
99.9 |
19 |
Fold:PLP-dependent transferase-like
Superfamily:PLP-dependent transferases
Family:AAT-like |
| 93 | c3dxvA_ |
|
not modelled |
99.9 |
12 |
PDB header:isomerase
Chain: A: PDB Molecule:alpha-amino-epsilon-caprolactam racemase;
PDBTitle: the crystal structure of alpha-amino-epsilon-caprolactam racemase from2 achromobacter obae
|
| 94 | c3ecdC_ |
|
not modelled |
99.9 |
16 |
PDB header:transferase
Chain: C: PDB Molecule:serine hydroxymethyltransferase 2;
PDBTitle: crystal structure of serine hydroxymethyltransferase from burkholderia2 pseudomallei
|
| 95 | c2hzpA_ |
|
not modelled |
99.9 |
14 |
PDB header:hydrolase
Chain: A: PDB Molecule:kynureninase;
PDBTitle: crystal structure of homo sapiens kynureninase
|
| 96 | d1dfoa_ |
|
not modelled |
99.9 |
13 |
Fold:PLP-dependent transferase-like
Superfamily:PLP-dependent transferases
Family:GABA-aminotransferase-like |
| 97 | c3h14A_ |
|
not modelled |
99.9 |
15 |
PDB header:transferase
Chain: A: PDB Molecule:aminotransferase, classes i and ii;
PDBTitle: crystal structure of a putative aminotransferase from silicibacter2 pomeroyi
|
| 98 | c3oksB_ |
|
not modelled |
99.9 |
13 |
PDB header:transferase
Chain: B: PDB Molecule:4-aminobutyrate transaminase;
PDBTitle: crystal structure of 4-aminobutyrate transaminase from mycobacterium2 smegmatis
|
| 99 | d1bw0a_ |
|
not modelled |
99.9 |
13 |
Fold:PLP-dependent transferase-like
Superfamily:PLP-dependent transferases
Family:AAT-like |
| 100 | c3hqtB_ |
|
not modelled |
99.9 |
13 |
PDB header:transferase
Chain: B: PDB Molecule:cai-1 autoinducer synthase;
PDBTitle: plp-dependent acyl-coa transferase cqsa
|
| 101 | c1c4kA_ |
|
not modelled |
99.9 |
14 |
PDB header:lyase
Chain: A: PDB Molecule:protein (ornithine decarboxylase);
PDBTitle: ornithine decarboxylase mutant (gly121tyr)
|
| 102 | d1sffa_ |
|
not modelled |
99.9 |
13 |
Fold:PLP-dependent transferase-like
Superfamily:PLP-dependent transferases
Family:GABA-aminotransferase-like |
| 103 | d1w7la_ |
|
not modelled |
99.9 |
15 |
Fold:PLP-dependent transferase-like
Superfamily:PLP-dependent transferases
Family:AAT-like |
| 104 | c3nraA_ |
|
not modelled |
99.9 |
18 |
PDB header:transferase
Chain: A: PDB Molecule:aspartate aminotransferase;
PDBTitle: crystal structure of an aspartate aminotransferase (yp_354942.1) from2 rhodobacter sphaeroides 2.4.1 at 2.15 a resolution
|
| 105 | d2cfba1 |
|
not modelled |
99.9 |
14 |
Fold:PLP-dependent transferase-like
Superfamily:PLP-dependent transferases
Family:GABA-aminotransferase-like |
| 106 | d1tpla_ |
|
not modelled |
99.9 |
10 |
Fold:PLP-dependent transferase-like
Superfamily:PLP-dependent transferases
Family:Beta-eliminating lyases |
| 107 | c3hl2D_ |
|
not modelled |
99.9 |
13 |
PDB header:transferase
Chain: D: PDB Molecule:o-phosphoseryl-trna(sec) selenium transferase;
PDBTitle: the crystal structure of the human sepsecs-trnasec complex
|
| 108 | c3dc1A_ |
|
not modelled |
99.9 |
15 |
PDB header:transferase
Chain: A: PDB Molecule:kynurenine/alpha-aminoadipate aminotransferase
PDBTitle: crystal structure of kynurenine aminotransferase ii complex with2 alpha-ketoglutarate
|
| 109 | d1vjoa_ |
|
not modelled |
99.9 |
16 |
Fold:PLP-dependent transferase-like
Superfamily:PLP-dependent transferases
Family:Cystathionine synthase-like |
| 110 | c3nnkC_ |
|
not modelled |
99.9 |
16 |
PDB header:transferase
Chain: C: PDB Molecule:ureidoglycine-glyoxylate aminotransferase;
PDBTitle: biochemical and structural characterization of a ureidoglycine2 aminotransferase in the klebsiella pneumoniae uric acid catabolic3 pathway
|
| 111 | c3f9tB_ |
|
not modelled |
99.9 |
14 |
PDB header:lyase
Chain: B: PDB Molecule:l-tyrosine decarboxylase mfna;
PDBTitle: crystal structure of l-tyrosine decarboxylase mfna (ec 4.1.1.25)2 (np_247014.1) from methanococcus jannaschii at 2.11 a resolution
|
| 112 | c3f0hA_ |
|
not modelled |
99.9 |
11 |
PDB header:transferase
Chain: A: PDB Molecule:aminotransferase;
PDBTitle: crystal structure of aminotransferase (rer070207000802) from2 eubacterium rectale at 1.70 a resolution
|
| 113 | d1js3a_ |
|
not modelled |
99.9 |
13 |
Fold:PLP-dependent transferase-like
Superfamily:PLP-dependent transferases
Family:Pyridoxal-dependent decarboxylase |
| 114 | c3fcrA_ |
|
not modelled |
99.9 |
12 |
PDB header:transferase
Chain: A: PDB Molecule:putative aminotransferase;
PDBTitle: crystal structure of putative aminotransferase (yp_614685.1) from2 silicibacter sp. tm1040 at 1.80 a resolution
|
| 115 | c3i5tB_ |
|
not modelled |
99.9 |
14 |
PDB header:transferase
Chain: B: PDB Molecule:aminotransferase;
PDBTitle: crystal structure of aminotransferase prk07036 from rhodobacter2 sphaeroides kd131
|
| 116 | d2v1pa1 |
|
not modelled |
99.9 |
16 |
Fold:PLP-dependent transferase-like
Superfamily:PLP-dependent transferases
Family:Beta-eliminating lyases |
| 117 | c3ffrA_ |
|
not modelled |
99.9 |
11 |
PDB header:transferase
Chain: A: PDB Molecule:phosphoserine aminotransferase serc;
PDBTitle: crystal structure of a phosphoserine aminotransferase serc (chu_0995)2 from cytophaga hutchinsonii atcc 33406 at 1.75 a resolution
|
| 118 | d1j32a_ |
|
not modelled |
99.9 |
16 |
Fold:PLP-dependent transferase-like
Superfamily:PLP-dependent transferases
Family:AAT-like |
| 119 | c2o0rA_ |
|
not modelled |
99.9 |
17 |
PDB header:transferase
Chain: A: PDB Molecule:rv0858c (n-succinyldiaminopimelate aminotransferase);
PDBTitle: the three-dimensional structure of n-succinyldiaminopimelate2 aminotransferase from mycobacterium tuberculosis
|
| 120 | c2jisA_ |
|
not modelled |
99.9 |
12 |
PDB header:lyase
Chain: A: PDB Molecule:cysteine sulfinic acid decarboxylase;
PDBTitle: human cysteine sulfinic acid decarboxylase (csad) in2 complex with plp.
|