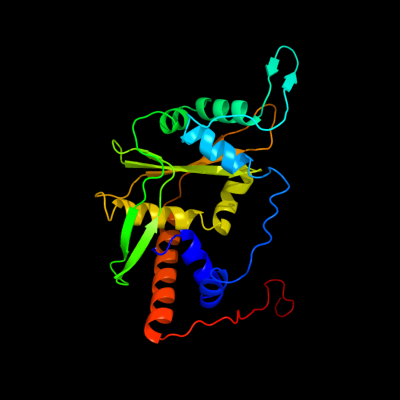
1 c3h4rA_
100.0
100
PDB header: hydrolaseChain: A: PDB Molecule: exodeoxyribonuclease 8;PDBTitle: crystal structure of e. coli rece exonuclease
2 c3l0aA_
100.0
19
PDB header: hydrolaseChain: A: PDB Molecule: putative exonuclease;PDBTitle: crystal structure of putative exonuclease (rer070207002219) from2 eubacterium rectale at 2.19 a resolution
3 c1w36E_
97.6
21
PDB header: recombinationChain: E: PDB Molecule: exodeoxyribonuclease v beta chain;PDBTitle: recbcd:dna complex
4 d1w36b3
96.7
19
Fold: Restriction endonuclease-likeSuperfamily: Restriction endonuclease-likeFamily: Exodeoxyribonuclease V beta chain (RecB), C-terminal domain5 d1rhoa_
54.1
50
Fold: Immunoglobulin-like beta-sandwichSuperfamily: E set domainsFamily: RhoGDI-like6 d2fhzb1
48.6
56
Fold: Colicin D/E5 nuclease domainSuperfamily: Colicin D/E5 nuclease domainFamily: Colicin E5 nuclease domain7 c2nuhA_
37.6
18
PDB header: unknown functionChain: A: PDB Molecule: periplasmic divalent cation tolerance protein;PDBTitle: crystal structure of cuta from the phytopathgen bacterium xylella2 fastidiosa
8 c2v43A_
36.2
36
PDB header: regulatorChain: A: PDB Molecule: sigma-e factor regulatory protein rseb;PDBTitle: crystal structure of rseb: a sensor for periplasmic stress2 response in e. coli
9 d1nzaa_
34.3
18
Fold: Ferredoxin-likeSuperfamily: GlnB-likeFamily: Divalent ion tolerance proteins CutA (CutA1)10 d1naqa_
33.2
26
Fold: Ferredoxin-likeSuperfamily: GlnB-likeFamily: Divalent ion tolerance proteins CutA (CutA1)11 c2zomC_
32.2
18
PDB header: unknown functionChain: C: PDB Molecule: protein cuta, chloroplast, putative, expressed;PDBTitle: crystal structure of cuta1 from oryza sativa
12 c3ahpA_
31.1
14
PDB header: electron transportChain: A: PDB Molecule: cuta1;PDBTitle: crystal structure of stable protein, cuta1, from a psychrotrophic2 bacterium shewanella sp. sib1
13 d1k8ta_
30.7
16
Fold: Adenylylcyclase toxin (the edema factor)Superfamily: Adenylylcyclase toxin (the edema factor)Family: Adenylylcyclase toxin (the edema factor)14 c3bb5B_
30.2
20
PDB header: unknown functionChain: B: PDB Molecule: stress responsive alpha-beta protein;PDBTitle: crystal structure of a dimeric ferredoxin-like protein of unknown2 function (jann_3925) from jannaschia sp. ccs1 at 2.30 a resolution
15 d2zfha1
29.5
16
Fold: Ferredoxin-likeSuperfamily: GlnB-likeFamily: Divalent ion tolerance proteins CutA (CutA1)16 c1xk8A_
29.5
16
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: divalent cation tolerant protein cuta;PDBTitle: divalent cation tolerant protein cuta from homo sapiens2 o60888
17 c2zfhA_
29.1
16
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: cuta;PDBTitle: crystal structure of putative cuta1 from homo sapiens at 2.05a2 resolution
18 c3d0jA_
28.5
27
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: uncharacterized protein ca_c3497;PDBTitle: crystal structure of conserved protein of unknown function ca_c34972 from clostridium acetobutylicum atcc 824
19 c1fybA_
27.2
60
PDB header: hydrolase inhibitorChain: A: PDB Molecule: proteinase inhibitor;PDBTitle: solution structure of c1-t1, a two-domain proteinase2 inhibitor derived from the circular precursor protein na-3 propi from nicotiana alata
20 d1p1la_
25.2
19
Fold: Ferredoxin-likeSuperfamily: GlnB-likeFamily: Divalent ion tolerance proteins CutA (CutA1)21 d1vhfa_
not modelled
22.6
17
Fold: Ferredoxin-likeSuperfamily: GlnB-likeFamily: Divalent ion tolerance proteins CutA (CutA1)22 d1osce_
not modelled
21.9
15
Fold: Ferredoxin-likeSuperfamily: GlnB-likeFamily: Divalent ion tolerance proteins CutA (CutA1)23 d1ukua_
not modelled
20.5
17
Fold: Ferredoxin-likeSuperfamily: GlnB-likeFamily: Divalent ion tolerance proteins CutA (CutA1)24 d1kr4a_
not modelled
19.7
17
Fold: Ferredoxin-likeSuperfamily: GlnB-likeFamily: Divalent ion tolerance proteins CutA (CutA1)25 d3bgea1
not modelled
18.7
18
Fold: post-AAA+ oligomerization domain-likeSuperfamily: post-AAA+ oligomerization domain-likeFamily: MgsA/YrvN C-terminal domain-like26 d3ctda1
not modelled
18.2
21
Fold: post-AAA+ oligomerization domain-likeSuperfamily: post-AAA+ oligomerization domain-likeFamily: MgsA/YrvN C-terminal domain-like27 d1ajwa_
not modelled
18.2
40
Fold: Immunoglobulin-like beta-sandwichSuperfamily: E set domainsFamily: RhoGDI-like28 c3et6A_
not modelled
15.0
13
PDB header: lyaseChain: A: PDB Molecule: soluble guanylyl cyclase beta;PDBTitle: the crystal structure of the catalytic domain of a eukaryotic2 guanylate cyclase
29 d1gpja1
not modelled
15.0
14
Fold: Glutamyl tRNA-reductase dimerization domainSuperfamily: Glutamyl tRNA-reductase dimerization domainFamily: Glutamyl tRNA-reductase dimerization domain30 c3nj2B_
not modelled
14.9
27
PDB header: unknown functionChain: B: PDB Molecule: duf269-containing protein;PDBTitle: crystal structure of cce_0566 from the cyanobacterium cyanothece2 51142, a protein associated with nitrogen fixation from the duf2693 family
31 c3iyuY_
not modelled
14.4
14
PDB header: virusChain: Y: PDB Molecule: outer capsid protein vp4;PDBTitle: atomic model of an infectious rotavirus particle
32 c3g43F_
not modelled
13.6
30
PDB header: metal binding proteinChain: F: PDB Molecule: voltage-dependent l-type calcium channel subunitPDBTitle: crystal structure of the calmodulin-bound cav1.2 c-terminal2 regulatory domain dimer
33 c3kolA_
not modelled
13.2
44
PDB header: metal binding proteinChain: A: PDB Molecule: glyoxalase/bleomycin resistancePDBTitle: crystal structure of a glyoxalase/dioxygenase from nostoc2 punctiforme
34 d1pjua1
not modelled
12.9
50
Fold: Plant proteinase inhibitorsSuperfamily: Plant proteinase inhibitorsFamily: Plant proteinase inhibitors35 c1h4tD_
not modelled
12.6
24
PDB header: aminoacyl-trna synthetaseChain: D: PDB Molecule: prolyl-trna synthetase;PDBTitle: prolyl-trna synthetase from thermus thermophilus complexed2 with l-proline
36 c1wq6A_
not modelled
12.5
29
PDB header: oncoproteinChain: A: PDB Molecule: aml1-eto;PDBTitle: the tetramer structure of the nervy homolgy two (nhr2) domain of aml1-2 eto is critical for aml1-eto's activity
37 c2o61A_
not modelled
12.3
32
PDB header: transcription/dnaChain: A: PDB Molecule: transcription factor p65/interferon regulatory factorPDBTitle: crystal structure of nfkb, irf7, irf3 bound to the interferon-b2 enhancer
38 c2q2kA_
not modelled
12.0
9
PDB header: dna binding protein/dnaChain: A: PDB Molecule: hypothetical protein;PDBTitle: structure of nucleic-acid binding protein
39 d4sgbi_
not modelled
11.8
60
Fold: Plant proteinase inhibitorsSuperfamily: Plant proteinase inhibitorsFamily: Plant proteinase inhibitors40 d1nr3a_
not modelled
11.3
5
Fold: DNA-binding protein TfxSuperfamily: DNA-binding protein TfxFamily: DNA-binding protein Tfx41 c3mcrA_
not modelled
11.1
32
PDB header: oxidoreductaseChain: A: PDB Molecule: nadh dehydrogenase, subunit c;PDBTitle: crystal structure of nadh dehydrogenase subunit c (tfu_2693) from2 thermobifida fusca yx-er1 at 2.65 a resolution
42 c2qycA_
not modelled
10.8
24
PDB header: unknown functionChain: A: PDB Molecule: ferredoxin-like protein;PDBTitle: crystal structure of a dimeric ferredoxin-like protein (bb1511) from2 bordetella bronchiseptica rb50 at 1.90 a resolution
43 d1oyvi_
not modelled
10.7
50
Fold: Plant proteinase inhibitorsSuperfamily: Plant proteinase inhibitorsFamily: Plant proteinase inhibitors44 c2l1nA_
not modelled
10.7
17
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: uncharacterized protein;PDBTitle: solution nmr structure of the protein yp_399305.1
45 c3bn7A_
not modelled
10.7
16
PDB header: unknown functionChain: A: PDB Molecule: ferredoxin-like protein;PDBTitle: crystal structure of a dimeric ferredoxin-like protein (cc_2267) from2 caulobacter crescentus cb15 at 1.64 a resolution
46 d1k3ka_
not modelled
10.5
16
Fold: Toxins' membrane translocation domainsSuperfamily: Bcl-2 inhibitors of programmed cell deathFamily: Bcl-2 inhibitors of programmed cell death47 c2khrA_
not modelled
10.3
67
PDB header: biosynthetic proteinChain: A: PDB Molecule: protein mbth;PDBTitle: solution structure of rv2377c, a mbth-like protein from mycobacterium2 tuberculosis
48 c2xgfA_
not modelled
9.9
26
PDB header: viral proteinChain: A: PDB Molecule: long tail fiber protein p37;PDBTitle: structure of the bacteriophage t4 long tail fibre needle-2 shaped receptor-binding tip
49 c3nsjA_
not modelled
9.3
19
PDB header: immune systemChain: A: PDB Molecule: perforin-1;PDBTitle: the x-ray crystal structure of lymphocyte perforin
50 c1w36F_
not modelled
9.1
12
PDB header: recombinationChain: F: PDB Molecule: exodeoxyribonuclease v gamma chain;PDBTitle: recbcd:dna complex
51 d1azsb_
not modelled
8.8
14
Fold: Ferredoxin-likeSuperfamily: Nucleotide cyclaseFamily: Adenylyl and guanylyl cyclase catalytic domain52 c2oevA_
not modelled
8.3
15
PDB header: protein transportChain: A: PDB Molecule: programmed cell death 6-interacting protein;PDBTitle: crystal structure of alix/aip1
53 d1leha2
not modelled
8.3
20
Fold: Aminoacid dehydrogenase-like, N-terminal domainSuperfamily: Aminoacid dehydrogenase-like, N-terminal domainFamily: Aminoacid dehydrogenases54 d2f23a1
not modelled
8.2
22
Fold: Long alpha-hairpinSuperfamily: GreA transcript cleavage protein, N-terminal domainFamily: GreA transcript cleavage protein, N-terminal domain55 c2rmsB_
not modelled
8.2
32
PDB header: transcriptionChain: B: PDB Molecule: msin3a-binding protein;PDBTitle: solution structure of the msin3a pah1-sap25 sid complex
56 c2bf9A_
not modelled
7.9
17
PDB header: hormoneChain: A: PDB Molecule: pancreatic hormone;PDBTitle: anisotropic refinement of avian (turkey) pancreatic2 polypeptide at 0.99 angstroms resolution.
57 d1fyba1
not modelled
7.7
60
Fold: Plant proteinase inhibitorsSuperfamily: Plant proteinase inhibitorsFamily: Plant proteinase inhibitors58 d2ftxa1
not modelled
7.5
53
Fold: Kinetochore globular domain-likeSuperfamily: Kinetochore globular domainFamily: Spc25-like59 c2hr5B_
not modelled
7.3
20
PDB header: metal binding proteinChain: B: PDB Molecule: rubrerythrin;PDBTitle: pf1283- rubrerythrin from pyrococcus furiosus iron bound form
60 c3ctdB_
not modelled
7.2
21
PDB header: structural genomics, unknown functionChain: B: PDB Molecule: putative atpase, aaa family;PDBTitle: crystal structure of a putative aaa family atpase from2 prochlorococcus marinus subsp. pastoris
61 d1g7oa1
not modelled
7.1
21
Fold: GST C-terminal domain-likeSuperfamily: GST C-terminal domain-likeFamily: Glutathione S-transferase (GST), C-terminal domain62 c1tz5A_
not modelled
7.1
17
PDB header: hormone/growth factorChain: A: PDB Molecule: chimera of pancreatic hormone and neuropeptide y;PDBTitle: [pnpy19-23]-hpp bound to dpc micelles
63 d1tzfa_
not modelled
7.0
14
Fold: Nucleotide-diphospho-sugar transferasesSuperfamily: Nucleotide-diphospho-sugar transferasesFamily: Cytidylytransferase64 c3u7vA_
not modelled
7.0
36
PDB header: hydrolaseChain: A: PDB Molecule: beta-galactosidase;PDBTitle: the structure of a putative beta-galactosidase from caulobacter2 crescentus cb15.
65 c2xnsC_
not modelled
6.8
71
PDB header: hydrolase/peptideChain: C: PDB Molecule: regulator of g-protein signaling 14;PDBTitle: crystal structure of human g alpha i1 bound to a designed helical2 peptide derived from the goloco motif of rgs14
66 c1hk8A_
not modelled
6.6
44
PDB header: oxidoreductaseChain: A: PDB Molecule: anaerobic ribonucleotide-triphosphate reductase;PDBTitle: structural basis for allosteric substrate specificity2 regulation in class iii ribonucleotide reductases:3 nrdd in complex with dgtp
67 d1hk8a_
not modelled
6.6
44
Fold: PFL-like glycyl radical enzymesSuperfamily: PFL-like glycyl radical enzymesFamily: Class III anaerobic ribonucleotide reductase NRDD subunit68 d1xpma2
not modelled
6.5
14
Fold: Thiolase-likeSuperfamily: Thiolase-likeFamily: Chalcone synthase-like69 d3erja1
not modelled
6.5
22
Fold: Peptidyl-tRNA hydrolase IISuperfamily: Peptidyl-tRNA hydrolase IIFamily: Peptidyl-tRNA hydrolase II70 c2hfvA_
not modelled
6.3
33
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: hypothetical protein rpa1041;PDBTitle: solution nmr structure of protein rpa1041 from pseudomonas2 aeruginosa. northeast structural genomics consortium3 target pat90.
71 c3f4yF_
not modelled
6.3
26
PDB header: viral proteinChain: F: PDB Molecule: mutant peptide derived from hiv gp41 chr domain;PDBTitle: hiv gp41 six-helix bundle containing a mutant chr alpha-2 peptide sequence
72 d1f06a2
not modelled
6.2
23
Fold: FwdE/GAPDH domain-likeSuperfamily: Glyceraldehyde-3-phosphate dehydrogenase-like, C-terminal domainFamily: Dihydrodipicolinate reductase-like73 d1m8aa_
not modelled
6.1
45
Fold: IL8-likeSuperfamily: Interleukin 8-like chemokinesFamily: Interleukin 8-like chemokines74 c2rbcA_
not modelled
6.1
8
PDB header: transferaseChain: A: PDB Molecule: sugar kinase;PDBTitle: crystal structure of a putative ribokinase from agrobacterium2 tumefaciens
75 d1tiha_
not modelled
6.1
60
Fold: Plant proteinase inhibitorsSuperfamily: Plant proteinase inhibitorsFamily: Plant proteinase inhibitors76 c1xtyD_
not modelled
6.0
18
PDB header: hydrolaseChain: D: PDB Molecule: peptidyl-trna hydrolase;PDBTitle: crystal structure of sulfolobus solfataricus peptidyl-trna2 hydrolase
77 d1q7sa_
not modelled
5.9
20
Fold: Peptidyl-tRNA hydrolase IISuperfamily: Peptidyl-tRNA hydrolase IIFamily: Peptidyl-tRNA hydrolase II78 d1dl6a_
not modelled
5.9
29
Fold: Rubredoxin-likeSuperfamily: Zinc beta-ribbonFamily: Transcriptional factor domain79 c3g7pA_
not modelled
5.9
21
PDB header: unknown functionChain: A: PDB Molecule: nitrogen fixation protein;PDBTitle: crystal structure of a nifx-associated protein of unknown function2 (afe_1514) from acidithiobacillus ferrooxidans atcc at 2.00 a3 resolution
80 c2wx6B_
not modelled
5.8
44
PDB header: oxidoreductaseChain: B: PDB Molecule: peroxidase ycdb;PDBTitle: x-ray crystallographic structure of e. coli apo-efeb
81 c2dezA_
not modelled
5.8
31
PDB header: neuropeptideChain: A: PDB Molecule: peptide yy;PDBTitle: structure of human pyy
82 c3be3A_
not modelled
5.8
50
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: uncharacterized protein;PDBTitle: crystal structure of a protein belonging to pfam duf16532 from bordetella bronchiseptica
83 c1ronA_
not modelled
5.7
13
PDB header: neuropeptideChain: A: PDB Molecule: neuropeptide y;PDBTitle: nmr solution structure of human neuropeptide y
84 d2gvka1
not modelled
5.7
50
Fold: Ferredoxin-likeSuperfamily: Dimeric alpha+beta barrelFamily: Dyp-type peroxidase-like85 d1rlka_
not modelled
5.6
12
Fold: Peptidyl-tRNA hydrolase IISuperfamily: Peptidyl-tRNA hydrolase IIFamily: Peptidyl-tRNA hydrolase II86 c2yy8B_
not modelled
5.6
29
PDB header: transferaseChain: B: PDB Molecule: upf0106 protein ph0461;PDBTitle: crystal structure of archaeal trna-methylase for position2 56 (atrm56) from pyrococcus horikoshii, complexed with s-3 adenosyl-l-methionine
87 d1q4ra_
not modelled
5.5
19
Fold: Ferredoxin-likeSuperfamily: Dimeric alpha+beta barrelFamily: Plant stress-induced protein88 d2o3aa1
not modelled
5.5
29
Fold: alpha/beta knotSuperfamily: alpha/beta knotFamily: AF0751-like89 d2i2la1
not modelled
5.4
24
Fold: YopX-likeSuperfamily: YopX-likeFamily: YopX-like90 d1nrja_
not modelled
5.4
41
Fold: Profilin-likeSuperfamily: SNARE-likeFamily: SRP alpha N-terminal domain-like91 c3c18B_
not modelled
5.4
9
PDB header: transferaseChain: B: PDB Molecule: nucleotidyltransferase-like protein;PDBTitle: crystal structure of nucleotidyltransferase-like protein2 (zp_00538802.1) from exiguobacterium sibiricum 255-15 at 1.90 a3 resolution
92 c1jsuC_
not modelled
5.4
35
PDB header: complex (transferase/cyclin/inhibitor)Chain: C: PDB Molecule: p27;PDBTitle: p27(kip1)/cyclin a/cdk2 complex
93 c2om2B_
not modelled
5.4
86
PDB header: signaling proteinChain: B: PDB Molecule: regulator of g-protein signalling 14 golocoPDBTitle: crystal structure of human g[alpha]i1 bound to the goloco2 motif of rgs14
94 d1wz3a1
not modelled
5.4
18
Fold: beta-Grasp (ubiquitin-like)Superfamily: Ubiquitin-likeFamily: APG12-like95 d2o8sa1
not modelled
5.3
21
Fold: AGR C 984p-likeSuperfamily: AGR C 984p-likeFamily: AGR C 984p-like96 c2rp4C_
not modelled
5.3
24
PDB header: transcriptionChain: C: PDB Molecule: transcription factor p53;PDBTitle: solution structure of the oligomerization domain in dmp53
97 c2d3kA_
not modelled
5.3
24
PDB header: hydrolaseChain: A: PDB Molecule: peptidyl-trna hydrolase;PDBTitle: structural study on project id ph1539 from pyrococcus2 horikoshii ot3
98 d1tr0a_
not modelled
5.3
19
Fold: Ferredoxin-likeSuperfamily: Dimeric alpha+beta barrelFamily: Plant stress-induced protein99 c2l4nA_
not modelled
5.2
36
PDB header: cytokineChain: A: PDB Molecule: c-c motif chemokine 21;PDBTitle: solution structure of the chemokine ccl21