| 1 |
|
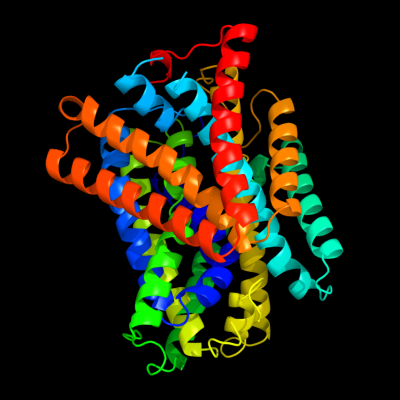
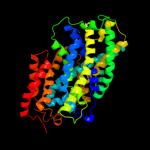
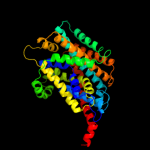
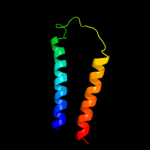
PDB 3gia chain A
Region: 20 - 469
Aligned: 426
Modelled: 426
Confidence: 100.0%
Identity: 12%
PDB header:transport protein
Chain: A: PDB Molecule:uncharacterized protein mj0609;
PDBTitle: crystal structure of apct transporter
Phyre2
| 2 |
|
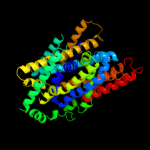
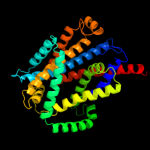
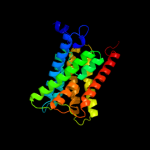
PDB 3lrc chain C
Region: 20 - 467
Aligned: 404
Modelled: 404
Confidence: 100.0%
Identity: 17%
PDB header:transport protein
Chain: C: PDB Molecule:arginine/agmatine antiporter;
PDBTitle: structure of e. coli adic (p1)
Phyre2
| 3 |
|
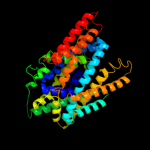
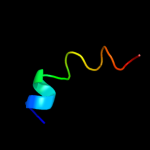
PDB 2jln chain A
Region: 6 - 470
Aligned: 447
Modelled: 451
Confidence: 100.0%
Identity: 10%
PDB header:membrane protein
Chain: A: PDB Molecule:mhp1;
PDBTitle: structure of mhp1, a nucleobase-cation-symport-1 family2 transporter
Phyre2
| 4 |
|
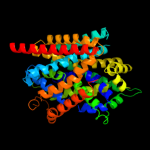
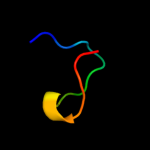
PDB 3dh4 chain A
Region: 3 - 470
Aligned: 452
Modelled: 436
Confidence: 99.6%
Identity: 10%
PDB header:transport protein
Chain: A: PDB Molecule:sodium/glucose cotransporter;
PDBTitle: crystal structure of sodium/sugar symporter with bound galactose from2 vibrio parahaemolyticus
Phyre2
| 5 |
|
PDB 2xq2 chain A
Region: 21 - 470
Aligned: 433
Modelled: 438
Confidence: 99.6%
Identity: 10%
PDB header:transport protein
Chain: A: PDB Molecule:sodium/glucose cotransporter;
PDBTitle: structure of the k294a mutant of vsglt
Phyre2
| 6 |
|
PDB 2a65 chain A domain 1
Region: 24 - 467
Aligned: 433
Modelled: 433
Confidence: 98.0%
Identity: 14%
Fold: SNF-like
Superfamily: SNF-like
Family: SNF-like
Phyre2
| 7 |
|
PDB 2w8a chain C
Region: 15 - 411
Aligned: 384
Modelled: 384
Confidence: 97.6%
Identity: 12%
PDB header:membrane protein
Chain: C: PDB Molecule:glycine betaine transporter betp;
PDBTitle: crystal structure of the sodium-coupled glycine betaine2 symporter betp from corynebacterium glutamicum with bound3 substrate
Phyre2
| 8 |
|
PDB 3hfx chain A
Region: 57 - 404
Aligned: 340
Modelled: 348
Confidence: 96.1%
Identity: 11%
PDB header:transport protein
Chain: A: PDB Molecule:l-carnitine/gamma-butyrobetaine antiporter;
PDBTitle: crystal structure of carnitine transporter
Phyre2
| 9 |
|
PDB 2l2t chain A
Region: 442 - 470
Aligned: 29
Modelled: 29
Confidence: 32.1%
Identity: 7%
PDB header:membrane protein
Chain: A: PDB Molecule:receptor tyrosine-protein kinase erbb-4;
PDBTitle: solution nmr structure of the erbb4 dimeric membrane domain
Phyre2
| 10 |
|
PDB 2jwa chain A
Region: 436 - 470
Aligned: 35
Modelled: 35
Confidence: 29.3%
Identity: 23%
PDB header:transferase
Chain: A: PDB Molecule:receptor tyrosine-protein kinase erbb-2;
PDBTitle: erbb2 transmembrane segment dimer spatial structure
Phyre2
| 11 |
|
PDB 1fft chain B domain 2
Region: 369 - 434
Aligned: 66
Modelled: 66
Confidence: 27.5%
Identity: 5%
Fold: Transmembrane helix hairpin
Superfamily: Cytochrome c oxidase subunit II-like, transmembrane region
Family: Cytochrome c oxidase subunit II-like, transmembrane region
Phyre2
| 12 |
|
PDB 1m57 chain H
Region: 370 - 434
Aligned: 65
Modelled: 65
Confidence: 18.6%
Identity: 17%
PDB header:oxidoreductase
Chain: H: PDB Molecule:cytochrome c oxidase;
PDBTitle: structure of cytochrome c oxidase from rhodobacter2 sphaeroides (eq(i-286) mutant))
Phyre2
| 13 |
|
PDB 2bbj chain B
Region: 412 - 463
Aligned: 52
Modelled: 52
Confidence: 12.6%
Identity: 13%
PDB header:metal transport/membrane protein
Chain: B: PDB Molecule:divalent cation transport-related protein;
PDBTitle: crystal structure of the cora mg2+ transporter
Phyre2
| 14 |
|
PDB 1ar1 chain B
Region: 372 - 434
Aligned: 63
Modelled: 63
Confidence: 9.8%
Identity: 10%
PDB header:complex (oxidoreductase/antibody)
Chain: B: PDB Molecule:cytochrome c oxidase;
PDBTitle: structure at 2.7 angstrom resolution of the paracoccus2 denitrificans two-subunit cytochrome c oxidase complexed3 with an antibody fv fragment
Phyre2
| 15 |
|
PDB 1qle chain B
Region: 372 - 434
Aligned: 63
Modelled: 63
Confidence: 9.8%
Identity: 10%
PDB header:oxidoreductase/immune system
Chain: B: PDB Molecule:cytochrome c oxidase polypeptide ii;
PDBTitle: cryo-structure of the paracoccus denitrificans four-subunit2 cytochrome c oxidase in the completely oxidized state3 complexed with an antibody fv fragment
Phyre2
| 16 |
|
PDB 2w2e chain A
Region: 3 - 241
Aligned: 239
Modelled: 239
Confidence: 8.5%
Identity: 11%
PDB header:membrane protein
Chain: A: PDB Molecule:aquaporin;
PDBTitle: 1.15 angstrom crystal structure of p.pastoris aquaporin,2 aqy1, in a closed conformation at ph 3.5
Phyre2
| 17 |
|
PDB 2rm9 chain A
Region: 2 - 19
Aligned: 18
Modelled: 18
Confidence: 8.1%
Identity: 17%
PDB header:neuropeptide
Chain: A: PDB Molecule:astressin2b;
PDBTitle: astressin2b
Phyre2
| 18 |
|
PDB 4a19 chain X
Region: 7 - 23
Aligned: 17
Modelled: 17
Confidence: 7.9%
Identity: 24%
PDB header:ribosome
Chain: X: PDB Molecule:rpl18a;
PDBTitle: t.thermophila 60s ribosomal subunit in complex with2 initiation factor 6. this file contains 26s rrna and3 proteins of molecule 2.
Phyre2
| 19 |
|
PDB 3ehb chain B domain 2
Region: 369 - 434
Aligned: 66
Modelled: 66
Confidence: 6.4%
Identity: 9%
Fold: Transmembrane helix hairpin
Superfamily: Cytochrome c oxidase subunit II-like, transmembrane region
Family: Cytochrome c oxidase subunit II-like, transmembrane region
Phyre2
| 20 |
|
PDB 3rko chain A
Region: 381 - 469
Aligned: 88
Modelled: 89
Confidence: 6.4%
Identity: 9%
PDB header:oxidoreductase
Chain: A: PDB Molecule:nadh-quinone oxidoreductase subunit a;
PDBTitle: crystal structure of the membrane domain of respiratory complex i from2 e. coli at 3.0 angstrom resolution
Phyre2
| 21 |
|
| 22 |
|
| 23 |
|