| 1 |
|
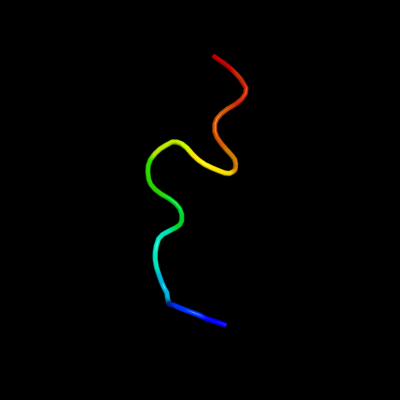
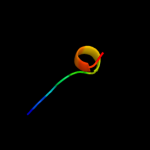
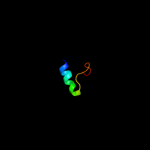
PDB 2k2w chain A
Region: 26 - 37
Aligned: 12
Modelled: 12
Confidence: 33.7%
Identity: 50%
PDB header:cell cycle
Chain: A: PDB Molecule:recombination and dna repair protein;
PDBTitle: second brct domain of nbs1
Phyre2
| 2 |
|
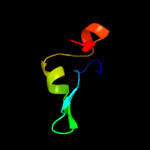
PDB 1twf chain J
Region: 6 - 52
Aligned: 33
Modelled: 33
Confidence: 17.2%
Identity: 36%
Fold: DNA/RNA-binding 3-helical bundle
Superfamily: RNA polymerase subunit RPB10
Family: RNA polymerase subunit RPB10
Phyre2
| 3 |
|
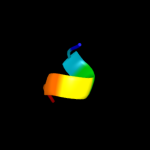
PDB 3czb chain A
Region: 19 - 33
Aligned: 15
Modelled: 15
Confidence: 16.0%
Identity: 20%
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:putative transglycosylase;
PDBTitle: crystal structure of putative transglycosylase from caulobacter2 crescentus
Phyre2
| 4 |
|
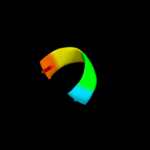
PDB 2pmz chain N
Region: 6 - 52
Aligned: 32
Modelled: 32
Confidence: 13.0%
Identity: 41%
PDB header:translation, transferase
Chain: N: PDB Molecule:dna-directed rna polymerase subunit n;
PDBTitle: archaeal rna polymerase from sulfolobus solfataricus
Phyre2
| 5 |
|
PDB 2pnw chain A domain 1
Region: 19 - 33
Aligned: 15
Modelled: 15
Confidence: 12.9%
Identity: 40%
Fold: Double psi beta-barrel
Superfamily: Barwin-like endoglucanases
Family: MLTA-like
Phyre2
| 6 |
|
PDB 2g5d chain A domain 1
Region: 19 - 33
Aligned: 15
Modelled: 15
Confidence: 11.3%
Identity: 33%
Fold: Double psi beta-barrel
Superfamily: Barwin-like endoglucanases
Family: MLTA-like
Phyre2
| 7 |
|
PDB 2pi8 chain A domain 1
Region: 7 - 33
Aligned: 27
Modelled: 26
Confidence: 10.7%
Identity: 26%
Fold: Double psi beta-barrel
Superfamily: Barwin-like endoglucanases
Family: MLTA-like
Phyre2
| 8 |
|
PDB 2ae0 chain X domain 1
Region: 19 - 33
Aligned: 15
Modelled: 15
Confidence: 10.3%
Identity: 40%
Fold: Double psi beta-barrel
Superfamily: Barwin-like endoglucanases
Family: MLTA-like
Phyre2
| 9 |
|
PDB 3cqy chain A
Region: 38 - 61
Aligned: 24
Modelled: 24
Confidence: 8.9%
Identity: 29%
PDB header:transferase
Chain: A: PDB Molecule:anhydro-n-acetylmuramic acid kinase;
PDBTitle: crystal structure of a functionally unknown protein (so_1313) from2 shewanella oneidensis mr-1
Phyre2
| 10 |
|
PDB 1niy chain A
Region: 31 - 43
Aligned: 13
Modelled: 13
Confidence: 8.6%
Identity: 54%
Fold: Knottins (small inhibitors, toxins, lectins)
Superfamily: omega toxin-like
Family: Spider toxins
Phyre2
| 11 |
|
PDB 1mb6 chain A
Region: 31 - 43
Aligned: 13
Modelled: 13
Confidence: 8.3%
Identity: 46%
Fold: Knottins (small inhibitors, toxins, lectins)
Superfamily: omega toxin-like
Family: Spider toxins
Phyre2
| 12 |
|
PDB 1b8t chain A domain 4
Region: 82 - 88
Aligned: 7
Modelled: 7
Confidence: 7.5%
Identity: 71%
Fold: Glucocorticoid receptor-like (DNA-binding domain)
Superfamily: Glucocorticoid receptor-like (DNA-binding domain)
Family: LIM domain
Phyre2
| 13 |
|
PDB 1b8t chain A domain 2
Region: 82 - 88
Aligned: 7
Modelled: 7
Confidence: 7.5%
Identity: 71%
Fold: Glucocorticoid receptor-like (DNA-binding domain)
Superfamily: Glucocorticoid receptor-like (DNA-binding domain)
Family: LIM domain
Phyre2
| 14 |
|
PDB 2djf chain A domain 1
Region: 38 - 47
Aligned: 10
Modelled: 10
Confidence: 7.1%
Identity: 40%
Fold: Streptavidin-like
Superfamily: Dipeptidyl peptidase I (cathepsin C), exclusion domain
Family: Dipeptidyl peptidase I (cathepsin C), exclusion domain
Phyre2
| 15 |
|
PDB 1mwp chain A
Region: 6 - 43
Aligned: 36
Modelled: 38
Confidence: 7.0%
Identity: 25%
Fold: SRCR-like
Superfamily: A heparin-binding domain
Family: A heparin-binding domain
Phyre2
| 16 |
|
PDB 1n93 chain X
Region: 47 - 52
Aligned: 6
Modelled: 6
Confidence: 6.8%
Identity: 50%
PDB header:viral protein
Chain: X: PDB Molecule:p40 nucleoprotein;
PDBTitle: crystal structure of the borna disease virus nucleoprotein
Phyre2
| 17 |
|
PDB 1n93 chain X
Region: 47 - 52
Aligned: 6
Modelled: 6
Confidence: 6.8%
Identity: 50%
Fold: P40 nucleoprotein
Superfamily: P40 nucleoprotein
Family: P40 nucleoprotein
Phyre2
| 18 |
|
PDB 1jxc chain A
Region: 20 - 29
Aligned: 10
Modelled: 10
Confidence: 6.7%
Identity: 50%
Fold: Knottins (small inhibitors, toxins, lectins)
Superfamily: Scorpion toxin-like
Family: Plant defensins
Phyre2
| 19 |
|
PDB 1jqp chain A domain 1
Region: 38 - 47
Aligned: 10
Modelled: 10
Confidence: 6.4%
Identity: 40%
Fold: Streptavidin-like
Superfamily: Dipeptidyl peptidase I (cathepsin C), exclusion domain
Family: Dipeptidyl peptidase I (cathepsin C), exclusion domain
Phyre2
| 20 |
|
PDB 3qbw chain A
Region: 38 - 61
Aligned: 24
Modelled: 24
Confidence: 6.2%
Identity: 29%
PDB header:transferase
Chain: A: PDB Molecule:anhydro-n-acetylmuramic acid kinase;
PDBTitle: crystal structure of pseudomonas aeruginosa 1,6-anhydro-n-2 actetylmuramic acid kinase (anmk) bound to adenosine diphosphate
Phyre2
| 21 |
|
| 22 |
|
| 23 |
|