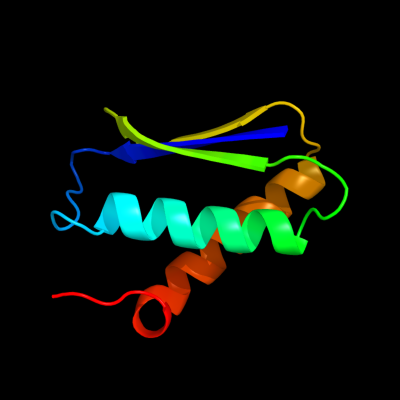
1 d1ghha_
100.0
100
Fold: DNA damage-inducible protein DinISuperfamily: DNA damage-inducible protein DinIFamily: DNA damage-inducible protein DinI2 c2j58G_
65.2
13
PDB header: membrane proteinChain: G: PDB Molecule: outer membrane lipoprotein wza;PDBTitle: the structure of wza
3 c2w8iG_
56.1
13
PDB header: membrane proteinChain: G: PDB Molecule: putative outer membrane lipoprotein wza;PDBTitle: crystal structure of wza24-345.
4 c3gr0D_
38.6
10
PDB header: membrane proteinChain: D: PDB Molecule: protein prgh;PDBTitle: periplasmic domain of the t3ss inner membrane protein prgh from2 s.typhimurium (fragment 170-362)
5 d1kn0a_
38.6
21
Fold: dsRBD-likeSuperfamily: dsRNA-binding domain-likeFamily: The homologous-pairing domain of Rad52 recombinase6 c3gr1A_
35.3
8
PDB header: membrane proteinChain: A: PDB Molecule: protein prgh;PDBTitle: periplamic domain of the t3ss inner membrane protein prgh2 from s.typhimurium (fragment 170-392)
7 d2akja1
23.9
19
Fold: Ferredoxin-likeSuperfamily: Nitrite/Sulfite reductase N-terminal domain-likeFamily: Duplicated SiR/NiR-like domains 1 and 38 d1ciya1
23.8
30
Fold: Galactose-binding domain-likeSuperfamily: Galactose-binding domain-likeFamily: delta-Endotoxin, C-terminal domain9 d1g5ma_
23.7
18
Fold: Toxins' membrane translocation domainsSuperfamily: Bcl-2 inhibitors of programmed cell deathFamily: Bcl-2 inhibitors of programmed cell death10 c2pfzA_
23.7
25
PDB header: transport proteinChain: A: PDB Molecule: putative exported protein;PDBTitle: crystal structure of dctp6, a bordetella pertussis2 extracytoplasmic solute receptor binding pyroglutamic acid
11 d1ji6a1
23.3
30
Fold: Galactose-binding domain-likeSuperfamily: Galactose-binding domain-likeFamily: delta-Endotoxin, C-terminal domain12 c1h2iG_
22.9
21
PDB header: dna-binding proteinChain: G: PDB Molecule: dna repair protein rad52 homolog;PDBTitle: human rad52 protein, n-terminal domain
13 d1zj8a1
21.4
9
Fold: Ferredoxin-likeSuperfamily: Nitrite/Sulfite reductase N-terminal domain-likeFamily: Duplicated SiR/NiR-like domains 1 and 314 c2zzxD_
20.4
17
PDB header: transport proteinChain: D: PDB Molecule: abc transporter, solute-binding protein;PDBTitle: crystal structure of a periplasmic substrate binding protein in2 complex with lactate
15 c2o90A_
19.8
13
PDB header: lyaseChain: A: PDB Molecule: dihydroneopterin aldolase;PDBTitle: atomic resolution crystal structure of e.coli2 dihydroneopterin aldolase in complex with neopterin
16 c2qfiB_
19.3
16
PDB header: transport proteinChain: B: PDB Molecule: ferrous-iron efflux pump fief;PDBTitle: structure of the zinc transporter yiip
17 d1aopa2
19.0
23
Fold: Ferredoxin-likeSuperfamily: Nitrite/Sulfite reductase N-terminal domain-likeFamily: Duplicated SiR/NiR-like domains 1 and 318 d1dlca1
18.1
13
Fold: Galactose-binding domain-likeSuperfamily: Galactose-binding domain-likeFamily: delta-Endotoxin, C-terminal domain19 c2z51A_
15.5
10
PDB header: metal transportChain: A: PDB Molecule: nifu-like protein 2, chloroplast;PDBTitle: crystal structure of arabidopsis cnfu involved in iron-2 sulfur cluster biosynthesis
20 c1usdA_
15.1
28
PDB header: signaling proteinChain: A: PDB Molecule: vasodilator-stimulated phosphoprotein;PDBTitle: human vasp tetramerisation domain l352m
21 c2xa0A_
not modelled
15.1
18
PDB header: apoptosisChain: A: PDB Molecule: apoptosis regulator bcl-2;PDBTitle: crystal structure of bcl-2 in complex with a bax bh32 peptide
22 d1ev0a_
not modelled
14.7
17
Fold: Cell division protein MinE topological specificity domainSuperfamily: Cell division protein MinE topological specificity domainFamily: Cell division protein MinE topological specificity domain23 c2v4jE_
not modelled
14.5
17
PDB header: oxidoreductaseChain: E: PDB Molecule: sulfite reductase, dissimilatory-type subunitPDBTitle: the crystal structure of desulfovibrio vulgaris2 dissimilatory sulfite reductase bound to dsrc provides3 novel insights into the mechanism of sulfate respiration
24 c2kz0A_
not modelled
12.7
27
PDB header: transcriptionChain: A: PDB Molecule: bola family protein;PDBTitle: solution structure of a bola protein (ech_0303) from ehrlichia2 chaffeensis. seattle structural genomics center for infectious3 disease target ehcha.10365.a
25 c2dbiA_
not modelled
12.3
20
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: hypothetical protein ybiu;PDBTitle: crystal structure of a hypothetical protein jw0805 from2 escherichia coli
26 c2cg8B_
not modelled
11.8
21
PDB header: lyase/transferaseChain: B: PDB Molecule: dihydroneopterin aldolase 6-hydroxymethyl-7,8-PDBTitle: the bifunctional dihydroneopterin aldolase 6-hydroxymethyl-2 7,8-dihydropterin synthase from streptococcus pneumoniae
27 c1zj8B_
not modelled
11.3
9
PDB header: oxidoreductaseChain: B: PDB Molecule: probable ferredoxin-dependent nitrite reductase nira;PDBTitle: structure of mycobacterium tuberculosis nira protein
28 c3u5gB_
not modelled
10.7
23
PDB header: ribosomeChain: B: PDB Molecule: 40s ribosomal protein s1-a;PDBTitle: the structure of the eukaryotic ribosome at 3.0 a resolution
29 d2csga1
not modelled
10.6
24
Fold: Double-stranded beta-helixSuperfamily: Clavaminate synthase-likeFamily: YbiU-like30 d1b9la_
not modelled
10.6
10
Fold: T-foldSuperfamily: Tetrahydrobiopterin biosynthesis enzymes-likeFamily: DHN aldolase/epimerase31 c3tr3A_
not modelled
10.2
10
PDB header: unknown functionChain: A: PDB Molecule: bola;PDBTitle: structure of a bola protein homologue from coxiella burnetii
32 c2w7aA_
not modelled
10.1
28
PDB header: rna-binding proteinChain: A: PDB Molecule: line-1 orf1p;PDBTitle: structure of the human line-1 orf1p central domain
33 c3d0wD_
not modelled
9.9
46
PDB header: structural genomics, unknown functionChain: D: PDB Molecule: yflh protein;PDBTitle: crystal structure of yflh protein from bacillus subtilis.2 northeast structural genomics consortium target sr326
34 c1xs3A_
not modelled
9.7
17
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: hypothetical protein xc975;PDBTitle: solution structure analysis of the xc975 protein
35 c2xzm4_
not modelled
9.2
14
PDB header: ribosomeChain: 4: PDB Molecule: 40s ribosomal protein s3a;PDBTitle: crystal structure of the eukaryotic 40s ribosomal2 subunit in complex with initiation factor 1. this file3 contains the 40s subunit and initiation factor for4 molecule 1
36 d1ekea_
not modelled
9.2
18
Fold: Ribonuclease H-like motifSuperfamily: Ribonuclease H-likeFamily: Ribonuclease H37 d1izna_
not modelled
8.9
22
Fold: Subunits of heterodimeric actin filament capping protein CapzSuperfamily: Subunits of heterodimeric actin filament capping protein CapzFamily: Capz alpha-1 subunit38 d1pv0a_
not modelled
8.5
29
Fold: Long alpha-hairpinSuperfamily: Sporulation inhibitor SdaFamily: Sporulation inhibitor Sda39 c2kxoA_
not modelled
8.4
7
PDB header: cell cycleChain: A: PDB Molecule: cell division topological specificity factor;PDBTitle: solution nmr structure of the cell division regulator mine protein2 from neisseria gonorrhoeae
40 c2yqrA_
not modelled
8.2
16
PDB header: rna binding proteinChain: A: PDB Molecule: kiaa0907 protein;PDBTitle: solution structure of the kh domain in kiaa0907 protein
41 c2jttD_
not modelled
8.0
29
PDB header: calcium binding protein/antitumor proteiChain: D: PDB Molecule: calcyclin-binding protein;PDBTitle: solution structure of calcium loaded s100a6 bound to c-2 terminal siah-1 interacting protein
42 d2qfia1
not modelled
8.0
15
Fold: Alpha-lytic protease prodomain-likeSuperfamily: Cation efflux protein cytoplasmic domain-likeFamily: Cation efflux protein cytoplasmic domain-like43 c1sqlN_
not modelled
7.8
12
PDB header: lyaseChain: N: PDB Molecule: dihydroneopterin aldolase;PDBTitle: crystal structure of 7,8-dihydroneopterin aldolase in2 complex with guanine
44 d1zbsa2
not modelled
7.7
17
Fold: Ribonuclease H-like motifSuperfamily: Actin-like ATPase domainFamily: BadF/BadG/BcrA/BcrD-like45 c2zztA_
not modelled
7.7
13
PDB header: transport proteinChain: A: PDB Molecule: putative uncharacterized protein;PDBTitle: crystal structure of the cytosolic domain of the cation2 diffusion facilitator family protein
46 c3r9jD_
not modelled
7.7
12
PDB header: cell cycle,hydrolase/cell cycleChain: D: PDB Molecule: cell division topological specificity factor;PDBTitle: 4.3a resolution structure of a mind-mine(i24n) protein complex
47 d2akja2
not modelled
7.7
16
Fold: Ferredoxin-likeSuperfamily: Nitrite/Sulfite reductase N-terminal domain-likeFamily: Duplicated SiR/NiR-like domains 1 and 348 d1a6qa1
not modelled
7.5
17
Fold: Another 3-helical bundleSuperfamily: Protein serine/threonine phosphatase 2C, C-terminal domainFamily: Protein serine/threonine phosphatase 2C, C-terminal domain49 d1k46a_
not modelled
7.4
15
Fold: YopH tyrosine phosphatase N-terminal domainSuperfamily: YopH tyrosine phosphatase N-terminal domainFamily: YopH tyrosine phosphatase N-terminal domain50 d2ponb1
not modelled
7.3
20
Fold: Toxins' membrane translocation domainsSuperfamily: Bcl-2 inhibitors of programmed cell deathFamily: Bcl-2 inhibitors of programmed cell death51 d1zj8a2
not modelled
6.9
18
Fold: Ferredoxin-likeSuperfamily: Nitrite/Sulfite reductase N-terminal domain-likeFamily: Duplicated SiR/NiR-like domains 1 and 352 c3c7bE_
not modelled
6.9
15
PDB header: oxidoreductaseChain: E: PDB Molecule: sulfite reductase, dissimilatory-type subunit beta;PDBTitle: structure of the dissimilatory sulfite reductase from archaeoglobus2 fulgidus
53 c3oxnD_
not modelled
6.9
14
PDB header: transcription regulatorChain: D: PDB Molecule: putative transcriptional regulator, lysr family;PDBTitle: the crystal structure of a putative transcriptional regulator from2 vibrio parahaemolyticus
54 c2veqA_
not modelled
6.7
35
PDB header: cell cycleChain: A: PDB Molecule: centromere dna-binding protein complex cbf3PDBTitle: insights into kinetochore-dna interactions from the2 structure of cep3p
55 c2kilA_
not modelled
6.6
6
PDB header: unknown functionChain: A: PDB Molecule: putative uncharacterized protein;PDBTitle: nmr structure of the h103g mutant so2144 h-nox domain from2 shewanella oneidensis in the fe(ii)co ligation state
56 d1d4ua1
not modelled
6.5
31
Fold: Putative DNA-binding domainSuperfamily: Putative DNA-binding domainFamily: DNA repair factor XPA DNA- and RPA-binding domain, C-terminal subdomain57 d3bypa1
not modelled
6.5
22
Fold: Alpha-lytic protease prodomain-likeSuperfamily: Cation efflux protein cytoplasmic domain-likeFamily: Cation efflux protein cytoplasmic domain-like58 c3fljA_
not modelled
6.3
24
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: uncharacterized protein conserved in bacteria with aPDBTitle: crystal structure of uncharacterized protein conserved in bacteria2 with a cystatin-like fold (yp_168589.1) from silicibacter pomeroyi3 dss-3 at 2.00 a resolution
59 d1wjwa_
not modelled
6.2
11
Fold: TBP-likeSuperfamily: Phosphoglucomutase, C-terminal domainFamily: Phosphoglucomutase, C-terminal domain60 d2esna2
not modelled
6.1
19
Fold: Periplasmic binding protein-like IISuperfamily: Periplasmic binding protein-like IIFamily: Phosphate binding protein-like61 c3dukD_
not modelled
6.1
26
PDB header: unknown functionChain: D: PDB Molecule: ntf2-like protein of unknown function;PDBTitle: crystal structure of a ntf2-like protein of unknown function2 (mfla_0564) from methylobacillus flagellatus kt at 2.200 a resolution
62 d1p90a_
not modelled
6.1
32
Fold: Ribonuclease H-like motifSuperfamily: Nitrogenase accessory factor-likeFamily: Nitrogenase accessory factor63 d2pa2a1
not modelled
6.0
17
Fold: alpha/beta-HammerheadSuperfamily: Ribosomal protein L16p/L10eFamily: Ribosomal protein L10e64 d1otka_
not modelled
5.7
16
Fold: Ferritin-likeSuperfamily: Ferritin-likeFamily: Ribonucleotide reductase-like65 c3c66B_
not modelled
5.7
22
PDB header: transferaseChain: B: PDB Molecule: poly(a) polymerase;PDBTitle: yeast poly(a) polymerase in complex with fip1 residues 80-105
66 c2akjA_
not modelled
5.6
15
PDB header: oxidoreductaseChain: A: PDB Molecule: ferredoxin--nitrite reductase, chloroplast;PDBTitle: structure of spinach nitrite reductase
67 d1fx0a1
not modelled
5.4
20
Fold: Left-handed superhelixSuperfamily: C-terminal domain of alpha and beta subunits of F1 ATP synthaseFamily: C-terminal domain of alpha and beta subunits of F1 ATP synthase68 d1pjqa3
not modelled
5.4
22
Fold: Siroheme synthase middle domains-likeSuperfamily: Siroheme synthase middle domains-likeFamily: Siroheme synthase middle domains-like69 d3c7bb2
not modelled
5.4
16
Fold: Ferredoxin-likeSuperfamily: Nitrite/Sulfite reductase N-terminal domain-likeFamily: DsrA/DsrB N-terminal-domain-like70 d1s3aa1
not modelled
5.3
18
Fold: Thioredoxin foldSuperfamily: Thioredoxin-likeFamily: Mitochondrial ribosomal protein L51/S25/CI-B8 domain71 c2l82A_
not modelled
5.2
33
PDB header: de novo proteinChain: A: PDB Molecule: designed protein or32;PDBTitle: solution nmr structure of de novo designed protein, p-loop ntpase2 fold, northeast structural genomics consortium target or32
72 d2ctja1
not modelled
5.1
18
Fold: Eukaryotic type KH-domain (KH-domain type I)Superfamily: Eukaryotic type KH-domain (KH-domain type I)Family: Eukaryotic type KH-domain (KH-domain type I)73 c2d4gA_
not modelled
5.1
16
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: hypothetical protein bsu11850;PDBTitle: structure of yjcg protein, a putative 2'-5' rna ligase from2 bacillus subtilis
74 d1q46a1
not modelled
5.1
22
Fold: SAM domain-likeSuperfamily: eIF2alpha middle domain-likeFamily: eIF2alpha middle domain-like75 d1q1oa_
not modelled
5.0
18
Fold: beta-Grasp (ubiquitin-like)Superfamily: CAD & PB1 domainsFamily: PB1 domain76 d1koya_
not modelled
5.0
38
Fold: C-terminal domain of DFF45/ICAD (DFF-C domain)Superfamily: C-terminal domain of DFF45/ICAD (DFF-C domain)Family: C-terminal domain of DFF45/ICAD (DFF-C domain)