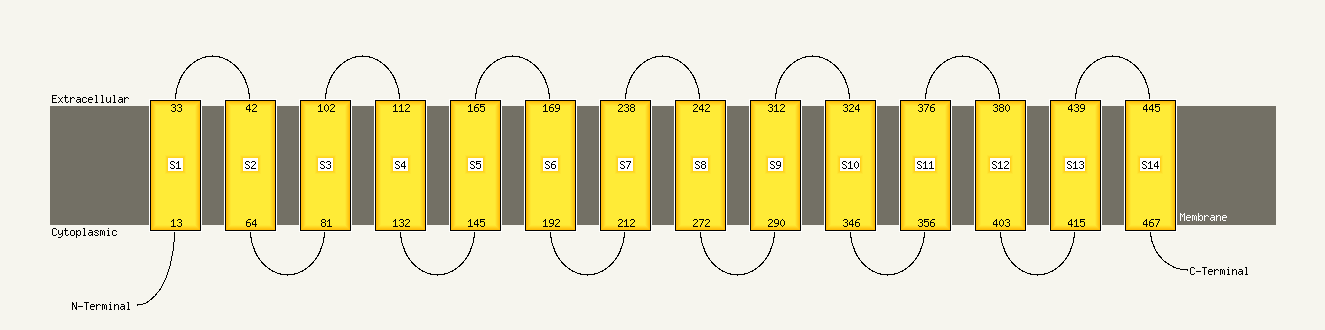
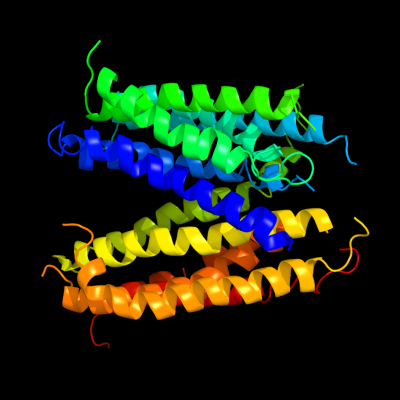
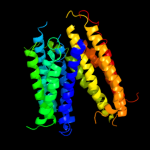
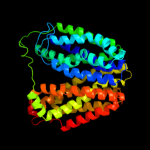
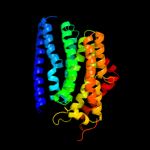
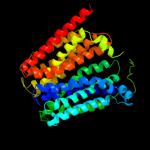
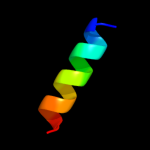
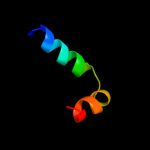
1 c3mkuA_
99.9
14
PDB header: transport proteinChain: A: PDB Molecule: multi antimicrobial extrusion protein (na(+)/drugPDBTitle: structure of a cation-bound multidrug and toxin compound extrusion2 (mate) transporter
2 d1pv7a_
65.5
10
Fold: MFS general substrate transporterSuperfamily: MFS general substrate transporterFamily: LacY-like proton/sugar symporter3 c3rkoN_
16.9
14
PDB header: oxidoreductaseChain: N: PDB Molecule: nadh-quinone oxidoreductase subunit n;PDBTitle: crystal structure of the membrane domain of respiratory complex i from2 e. coli at 3.0 angstrom resolution
4 d1pw4a_
12.4
7
Fold: MFS general substrate transporterSuperfamily: MFS general substrate transporterFamily: Glycerol-3-phosphate transporter5 d2iuba2
12.2
20
Fold: Transmembrane helix hairpinSuperfamily: Magnesium transport protein CorA, transmembrane regionFamily: Magnesium transport protein CorA, transmembrane region6 d1irxa1
11.8
21
Fold: An anticodon-binding domain of class I aminoacyl-tRNA synthetasesSuperfamily: An anticodon-binding domain of class I aminoacyl-tRNA synthetasesFamily: C-terminal domain of class I lysyl-tRNA synthetase7 c2jwaA_
11.2
17
PDB header: transferaseChain: A: PDB Molecule: receptor tyrosine-protein kinase erbb-2;PDBTitle: erbb2 transmembrane segment dimer spatial structure
8 c2pcoA_
10.9
6
PDB header: toxinChain: A: PDB Molecule: latarcin-1;PDBTitle: spatial structure and membrane permeabilization for2 latarcin-1, a spider antimicrobial peptide
9 c3iz5q_
10.8
20
PDB header: ribosomeChain: Q: PDB Molecule: 60s ribosomal protein l5 (l18p);PDBTitle: localization of the large subunit ribosomal proteins into a 5.5 a2 cryo-em map of triticum aestivum translating 80s ribosome
10 c3izrq_
10.8
20
PDB header: ribosomeChain: Q: PDB Molecule: 60s ribosomal protein l5 (l18p);PDBTitle: localization of the large subunit ribosomal proteins into a 5.5 a2 cryo-em map of triticum aestivum translating 80s ribosome
11 c2bbjB_
10.5
14
PDB header: metal transport/membrane proteinChain: B: PDB Molecule: divalent cation transport-related protein;PDBTitle: crystal structure of the cora mg2+ transporter
12 c2okrC_
10.0
30
PDB header: transferaseChain: C: PDB Molecule: map kinase-activated protein kinase 2;PDBTitle: crystal structure of the p38a-mapkap kinase 2 heterodimer
13 c2okrF_
10.0
30
PDB header: transferaseChain: F: PDB Molecule: map kinase-activated protein kinase 2;PDBTitle: crystal structure of the p38a-mapkap kinase 2 heterodimer
14 c2yukA_
9.9
15
PDB header: transferaseChain: A: PDB Molecule: myeloid/lymphoid or mixed-lineage leukemiaPDBTitle: solution structure of the hmg box of human myeloid/lymphoid2 or mixed-lineage leukemia protein 3 homolog
15 c3nd0A_
9.7
16
PDB header: transport proteinChain: A: PDB Molecule: sll0855 protein;PDBTitle: x-ray crystal structure of a slow cyanobacterial cl-/h+ antiporter
16 d1udxa3
9.3
16
Fold: Obg GTP-binding protein C-terminal domainSuperfamily: Obg GTP-binding protein C-terminal domainFamily: Obg GTP-binding protein C-terminal domain17 c2jo8B_
9.2
6
PDB header: transferaseChain: B: PDB Molecule: serine/threonine-protein kinase 4;PDBTitle: solution structure of c-terminal domain of human mammalian2 sterile 20-like kinase 1 (mst1)
18 c1b9uA_
9.0
29
PDB header: hydrolaseChain: A: PDB Molecule: protein (atp synthase);PDBTitle: membrane domain of the subunit b of the e.coli atp synthase
19 d1o82a_
9.0
12
Fold: Saposin-likeSuperfamily: Bacteriocin AS-48Family: Bacteriocin AS-4820 c1p84D_
8.9
9
PDB header: oxidoreductaseChain: D: PDB Molecule: cytochrome c1, heme protein;PDBTitle: hdbt inhibited yeast cytochrome bc1 complex
21 c1r6rA_
not modelled
8.7
15
PDB header: viral proteinChain: A: PDB Molecule: genome polyprotein;PDBTitle: solution structure of dengue virus capsid protein reveals a2 new fold
22 d1r6ra_
not modelled
8.7
15
Fold: Flavivirus capsid protein CSuperfamily: Flavivirus capsid protein CFamily: Flavivirus capsid protein C23 c1qvrB_
not modelled
8.7
23
PDB header: chaperoneChain: B: PDB Molecule: clpb protein;PDBTitle: crystal structure analysis of clpb
24 c3lmbA_
not modelled
8.4
21
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: uncharacterized protein;PDBTitle: the crystal structure of the protein olei01261 with unknown function2 from chlorobaculum tepidum tls
25 d2hkja2
not modelled
8.3
11
Fold: Ribosomal protein S5 domain 2-likeSuperfamily: Ribosomal protein S5 domain 2-likeFamily: DNA gyrase/MutL, second domain26 d1wwra1
not modelled
8.2
12
Fold: Cytidine deaminase-likeSuperfamily: Cytidine deaminase-likeFamily: Deoxycytidylate deaminase-like27 c2qdqA_
not modelled
8.2
9
PDB header: structural proteinChain: A: PDB Molecule: talin-1;PDBTitle: crystal structure of the talin dimerisation domain
28 d2q7ra1
not modelled
8.1
19
Fold: MAPEG domain-likeSuperfamily: MAPEG domain-likeFamily: MAPEG domain29 d2fyuk1
not modelled
8.1
16
Fold: Single transmembrane helixSuperfamily: Subunit XI (6.4 kDa protein) of cytochrome bc1 complex (Ubiquinol-cytochrome c reductase)Family: Subunit XI (6.4 kDa protein) of cytochrome bc1 complex (Ubiquinol-cytochrome c reductase)30 c2l16A_
not modelled
8.1
21
PDB header: protein transportChain: A: PDB Molecule: sec-independent protein translocase protein tatad;PDBTitle: solution structure of bacillus subtilits tatad protein in dpc micelles
31 d1sfka_
not modelled
8.0
17
Fold: Flavivirus capsid protein CSuperfamily: Flavivirus capsid protein CFamily: Flavivirus capsid protein C32 c3trrA_
not modelled
8.0
8
PDB header: isomeraseChain: A: PDB Molecule: probable enoyl-coa hydratase/isomerase;PDBTitle: crystal structure of a probable enoyl-coa hydratase/isomerase from2 mycobacterium abscessus
33 d3ehbb2
not modelled
8.0
9
Fold: Transmembrane helix hairpinSuperfamily: Cytochrome c oxidase subunit II-like, transmembrane regionFamily: Cytochrome c oxidase subunit II-like, transmembrane region34 c3cwbQ_
not modelled
8.0
6
PDB header: oxidoreductaseChain: Q: PDB Molecule: mitochondrial cytochrome c1, heme protein;PDBTitle: chicken cytochrome bc1 complex inhibited by an iodinated analogue of2 the polyketide crocacin-d
35 c3u5en_
not modelled
8.0
11
PDB header: ribosomeChain: N: PDB Molecule: 60s ribosomal protein l15-a;PDBTitle: the structure of the eukaryotic ribosome at 3.0 resolution
36 c3izcq_
not modelled
8.0
11
PDB header: ribosomeChain: Q: PDB Molecule: 60s ribosomal protein rpl5 (l18p);PDBTitle: localization of the large subunit ribosomal proteins into a 6.1 a2 cryo-em map of saccharomyces cerevisiae translating 80s ribosome
37 c3izsq_
not modelled
8.0
11
PDB header: ribosomeChain: Q: PDB Molecule: 60s ribosomal protein rpl5 (l18p);PDBTitle: localization of the large subunit ribosomal proteins into a 6.1 a2 cryo-em map of saccharomyces cerevisiae translating 80s ribosome
38 c3u5in_
not modelled
8.0
11
PDB header: ribosomeChain: N: PDB Molecule: 60s ribosomal protein l15-a;PDBTitle: the structure of the eukaryotic ribosome at 3.0 a resolution
39 d1j2jb_
not modelled
7.7
12
Fold: Spectrin repeat-likeSuperfamily: GAT-like domainFamily: GAT domain40 d2p7tc1
not modelled
7.7
17
Fold: Voltage-gated potassium channelsSuperfamily: Voltage-gated potassium channelsFamily: Voltage-gated potassium channels41 c2ww9B_
not modelled
7.7
8
PDB header: ribosomeChain: B: PDB Molecule: protein transport protein sss1;PDBTitle: cryo-em structure of the active yeast ssh1 complex bound to the2 yeast 80s ribosome
42 c1m2zB_
not modelled
7.6
44
PDB header: hormone/hormone activatorChain: B: PDB Molecule: nuclear receptor coactivator 2;PDBTitle: crystal structure of a dimer complex of the human2 glucocorticoid receptor ligand-binding domain bound to3 dexamethasone and a tif2 coactivator motif
43 c3qnqD_
not modelled
7.6
16
PDB header: membrane protein, transport proteinChain: D: PDB Molecule: pts system, cellobiose-specific iic component;PDBTitle: crystal structure of the transporter chbc, the iic component from the2 n,n'-diacetylchitobiose-specific phosphotransferase system
44 d1l0nk_
not modelled
7.6
15
Fold: Single transmembrane helixSuperfamily: Subunit XI (6.4 kDa protein) of cytochrome bc1 complex (Ubiquinol-cytochrome c reductase)Family: Subunit XI (6.4 kDa protein) of cytochrome bc1 complex (Ubiquinol-cytochrome c reductase)45 c3h81A_
not modelled
7.6
12
PDB header: lyaseChain: A: PDB Molecule: enoyl-coa hydratase echa8;PDBTitle: crystal structure of enoyl-coa hydratase from mycobacterium2 tuberculosis
46 c3pxiB_
not modelled
7.5
23
PDB header: protein bindingChain: B: PDB Molecule: negative regulator of genetic competence clpc/mecb;PDBTitle: structure of meca108:clpc
47 d1z3aa1
not modelled
7.5
11
Fold: Cytidine deaminase-likeSuperfamily: Cytidine deaminase-likeFamily: Deoxycytidylate deaminase-like48 d1e7la1
not modelled
7.4
24
Fold: LEM/SAP HeH motifSuperfamily: Recombination endonuclease VII, C-terminal and dimerization domainsFamily: Recombination endonuclease VII, C-terminal and dimerization domains49 c1m2zE_
not modelled
7.4
44
PDB header: hormone/hormone activatorChain: E: PDB Molecule: nuclear receptor coactivator 2;PDBTitle: crystal structure of a dimer complex of the human2 glucocorticoid receptor ligand-binding domain bound to3 dexamethasone and a tif2 coactivator motif
50 c3dh4A_
not modelled
7.4
12
PDB header: transport proteinChain: A: PDB Molecule: sodium/glucose cotransporter;PDBTitle: crystal structure of sodium/sugar symporter with bound galactose from2 vibrio parahaemolyticus
51 d1x6fa1
not modelled
7.3
32
Fold: beta-beta-alpha zinc fingersSuperfamily: beta-beta-alpha zinc fingersFamily: Classic zinc finger, C2H252 d1xqoa_
not modelled
7.2
6
Fold: DNA-glycosylaseSuperfamily: DNA-glycosylaseFamily: AgoG-like53 c2xq2A_
not modelled
7.2
11
PDB header: transport proteinChain: A: PDB Molecule: sodium/glucose cotransporter;PDBTitle: structure of the k294a mutant of vsglt
54 c1q2iA_
not modelled
7.1
10
PDB header: antitumor proteinChain: A: PDB Molecule: pnc27;PDBTitle: nmr solution structure of a peptide from the mdm-2 binding2 domain of the p53 protein that is selectively cytotoxic to3 cancer cells
55 c2z99A_
not modelled
7.1
13
PDB header: cell cycleChain: A: PDB Molecule: putative uncharacterized protein;PDBTitle: crystal structure of scpb from mycobacterium tuberculosis
56 c3fshC_
not modelled
7.1
12
PDB header: ligaseChain: C: PDB Molecule: autocrine motility factor receptor, isoform 2;PDBTitle: crystal structure of the ubiquitin conjugating enzyme2 ube2g2 bound to the g2br domain of ubiquitin ligase gp78
57 c3hinA_
not modelled
7.1
15
PDB header: lyaseChain: A: PDB Molecule: putative 3-hydroxybutyryl-coa dehydratase;PDBTitle: crystal structure of putative enoyl-coa hydratase from2 rhodopseudomonas palustris cga009
58 c1qcrD_
not modelled
7.0
6
PDB header: PDB COMPND: 59 c2ap7A_
not modelled
6.9
47
PDB header: antibioticChain: A: PDB Molecule: bombinin h2;PDBTitle: solution structure of bombinin h2 in dpc micelles
60 c3qooA_
not modelled
6.9
7
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: uncharacterized protein;PDBTitle: crystal structure of hot-dog-like taci_0573 protein from2 thermanaerovibrio acidaminovorans
61 d1cjya2
not modelled
6.9
0
Fold: FabD/lysophospholipase-likeSuperfamily: FabD/lysophospholipase-likeFamily: Lysophospholipase62 c2hn8A_
not modelled
6.9
17
PDB header: viral proteinChain: A: PDB Molecule: protein pb1-f2;PDBTitle: structural characterization and oligomerization of pb1-f2,2 a pro-apoptotic influenza a virus protein
63 c3jteA_
not modelled
6.8
16
PDB header: protein bindingChain: A: PDB Molecule: response regulator receiver protein;PDBTitle: crystal structure of response regulator receiver domain2 protein from clostridium thermocellum
64 c2hw5F_
not modelled
6.8
12
PDB header: lyaseChain: F: PDB Molecule: enoyl-coa hydratase;PDBTitle: the crystal structure of human enoyl-coenzyme a (coa) hydratase short2 chain 1, echs1
65 c3g64A_
not modelled
6.8
17
PDB header: lyaseChain: A: PDB Molecule: putative enoyl-coa hydratase;PDBTitle: crystal structure of putative enoyl-coa hydratase from streptomyces2 coelicolor a3(2)
66 c1yhnB_
not modelled
6.8
25
PDB header: protein transportChain: B: PDB Molecule: rab interacting lysosomal protein;PDBTitle: structure basis of rilp recruitment by rab7
67 d1jo5a_
not modelled
6.7
14
Fold: Light-harvesting complex subunitsSuperfamily: Light-harvesting complex subunitsFamily: Light-harvesting complex subunits68 c2qziA_
not modelled
6.7
11
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: uncharacterized protein;PDBTitle: the crystal structure of a conserved protein of unknown function from2 streptococcus thermophilus lmg 18311.
69 c3emlA_
not modelled
6.7
11
PDB header: membrane protein, receptorChain: A: PDB Molecule: human adenosine a2a receptor/t4 lysozyme chimera;PDBTitle: the 2.6 a crystal structure of a human a2a adenosine2 receptor bound to zm241385.
70 d1krra_
not modelled
6.7
30
Fold: Single-stranded left-handed beta-helixSuperfamily: Trimeric LpxA-like enzymesFamily: Galactoside acetyltransferase-like71 c2i88A_
not modelled
6.7
12
PDB header: membrane proteinChain: A: PDB Molecule: colicin-e1;PDBTitle: crystal structure of the channel-forming domain of colicin2 e1
72 c3a4cA_
not modelled
6.7
5
PDB header: cell cycle, replicationChain: A: PDB Molecule: dna replication factor cdt1;PDBTitle: crystal structure of cdt1 c terminal domain
73 c1sneA_
not modelled
6.6
100
PDB header: de novo proteinChain: A: PDB Molecule: tetrameric beta-beta-alpha mini-protein;PDBTitle: an oligomeric domain-swapped beta-beta-alpha mini-protein
74 c1sneB_
not modelled
6.6
100
PDB header: de novo proteinChain: B: PDB Molecule: tetrameric beta-beta-alpha mini-protein;PDBTitle: an oligomeric domain-swapped beta-beta-alpha mini-protein
75 d2diia1
not modelled
6.5
19
Fold: BSD domain-likeSuperfamily: BSD domain-likeFamily: BSD domain76 c2qnaB_
not modelled
6.5
28
PDB header: transport proteinChain: B: PDB Molecule: snurportin-1;PDBTitle: crystal structure of human importin-beta (127-876) in complex with the2 ibb-domain of snurportin1 (1-65)
77 c2g9pA_
not modelled
6.5
19
PDB header: antimicrobial proteinChain: A: PDB Molecule: antimicrobial peptide latarcin 2a;PDBTitle: nmr structure of a novel antimicrobial peptide, latarcin 2a,2 from spider (lachesana tarabaevi) venom
78 d2a5yb1
not modelled
6.5
33
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: CED-4 C-terminal domain-like79 c1irxA_
not modelled
6.4
21
PDB header: ligaseChain: A: PDB Molecule: lysyl-trna synthetase;PDBTitle: crystal structure of class i lysyl-trna synthetase
80 c1vdfB_
not modelled
6.4
14
PDB header: extracellular matrix proteinChain: B: PDB Molecule: cartilage oligomeric matrix protein;PDBTitle: assembly domain of cartilage oligomeric matrix protein
81 c2diiA_
not modelled
6.4
19
PDB header: transcriptionChain: A: PDB Molecule: tfiih basal transcription factor complex p62PDBTitle: solution structure of the bsd domain of human tfiih basal2 transcription factor complex p62 subunit
82 c1t6sB_
not modelled
6.4
22
PDB header: structural genomics, unknown functionChain: B: PDB Molecule: conserved hypothetical protein;PDBTitle: crystal structure of a conserved hypothetical protein from chlorobium2 tepidum
83 c2ap8A_
not modelled
6.4
43
PDB header: antibioticChain: A: PDB Molecule: bombinin h4;PDBTitle: solution structure of bombinin h4 in dpc micelles
84 d2o5ha1
not modelled
6.3
10
Fold: NMB0513-likeSuperfamily: NMB0513-likeFamily: NMB0513-like85 c3hrxD_
not modelled
6.3
16
PDB header: lyaseChain: D: PDB Molecule: probable enoyl-coa hydratase;PDBTitle: crystal structure of phenylacetic acid degradation protein paag
86 c3kikF_
not modelled
6.3
10
PDB header: transcriptionChain: F: PDB Molecule: saga-associated factor 11;PDBTitle: sgf11:sus1 complex
87 c3kikE_
not modelled
6.3
10
PDB header: transcriptionChain: E: PDB Molecule: saga-associated factor 11;PDBTitle: sgf11:sus1 complex
88 c2kncA_
not modelled
6.3
16
PDB header: cell adhesionChain: A: PDB Molecule: integrin alpha-iib;PDBTitle: platelet integrin alfaiib-beta3 transmembrane-cytoplasmic2 heterocomplex
89 c1jdmA_
not modelled
6.3
3
PDB header: membrane proteinChain: A: PDB Molecule: sarcolipin;PDBTitle: nmr structure of sarcolipin
90 c3kikH_
not modelled
6.2
10
PDB header: transcriptionChain: H: PDB Molecule: saga-associated factor 11;PDBTitle: sgf11:sus1 complex
91 d1unda_
not modelled
6.2
14
Fold: VHP, Villin headpiece domainSuperfamily: VHP, Villin headpiece domainFamily: VHP, Villin headpiece domain92 c2nx8A_
not modelled
6.2
11
PDB header: hydrolaseChain: A: PDB Molecule: trna-specific adenosine deaminase;PDBTitle: the crystal structure of the trna-specific adenosine deaminase from2 streptococcus pyogenes
93 d1wgma_
not modelled
6.2
13
Fold: RING/U-boxSuperfamily: RING/U-boxFamily: U-box94 c3kikG_
not modelled
6.2
10
PDB header: transcriptionChain: G: PDB Molecule: saga-associated factor 11;PDBTitle: sgf11:sus1 complex
95 c1zxaB_
not modelled
6.2
11
PDB header: transferaseChain: B: PDB Molecule: cgmp-dependent protein kinase 1, alpha isozyme;PDBTitle: solution structure of the coiled-coil domain of cgmp-2 dependent protein kinase ia
96 c2kesA_
not modelled
6.2
33
PDB header: protein bindingChain: A: PDB Molecule: synphilin-1;PDBTitle: solution structure of the coiled-coil domain of synphilin-1
97 c2ww9O_
not modelled
6.1
14
PDB header: ribosomeChain: O: PDB Molecule: 60s ribosomal protein l39;PDBTitle: cryo-em structure of the active yeast ssh1 complex bound to the2 yeast 80s ribosome
98 c3kjlF_
not modelled
6.1
10
PDB header: transcriptionChain: F: PDB Molecule: saga-associated factor 11;PDBTitle: sgf11:sus1 complex
99 c3zs9D_
not modelled
6.1
29
PDB header: hydrolase/transport proteinChain: D: PDB Molecule: golgi to er traffic protein 2;PDBTitle: s. cerevisiae get3-adp-alf4- complex with a cytosolic get2 fragment