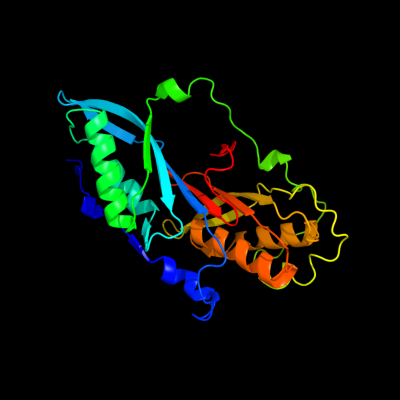
| 1 | c3bp1A_
|
|
 |
100.0 |
64 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:nadph-dependent 7-cyano-7-deazaguanine reductase;
PDBTitle: crystal structure of putative 7-cyano-7-deazaguanine2 reductase quef from vibrio cholerae o1 biovar eltor
|
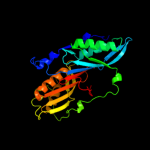
| 2 | c1wm9D_
|
|
 |
97.8 |
19 |
PDB header:hydrolase
Chain: D: PDB Molecule:gtp cyclohydrolase i;
PDBTitle: structure of gtp cyclohydrolase i from thermus thermophilus hb8
|
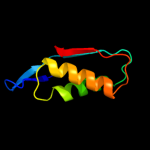
| 3 | d1wura1
|
|
 |
97.8 |
19 |
Fold:T-fold
Superfamily:Tetrahydrobiopterin biosynthesis enzymes-like
Family:GTP cyclohydrolase I |
| 4 | c1is7F_
|
|
 |
95.5 |
20 |
PDB header:hydrolase/protein binding
Chain: F: PDB Molecule:gtp cyclohydrolase i;
PDBTitle: crystal structure of rat gtpchi/gfrp stimulatory complex
|
| 5 | d1wpla_
|
|
 |
95.5 |
20 |
Fold:T-fold
Superfamily:Tetrahydrobiopterin biosynthesis enzymes-like
Family:GTP cyclohydrolase I |
| 6 | d1a8ra_
|
|
 |
95.2 |
19 |
Fold:T-fold
Superfamily:Tetrahydrobiopterin biosynthesis enzymes-like
Family:GTP cyclohydrolase I |
| 7 | c1rg9D_
|
|
 |
77.9 |
16 |
PDB header:transferase
Chain: D: PDB Molecule:s-adenosylmethionine synthetase;
PDBTitle: s-adenosylmethionine synthetase complexed with sam and ppnp
|
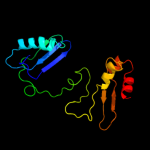
| 8 | c3imlB_
|
|
 |
60.2 |
18 |
PDB header:transferase
Chain: B: PDB Molecule:s-adenosylmethionine synthetase;
PDBTitle: crystal structure of s-adenosylmethionine synthetase from burkholderia2 pseudomallei
|
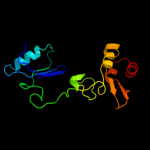
| 9 | c2obvA_
|
|
 |
52.0 |
16 |
PDB header:transferase
Chain: A: PDB Molecule:s-adenosylmethionine synthetase isoform type-1;
PDBTitle: crystal structure of the human s-adenosylmethionine synthetase 1 in2 complex with the product
|
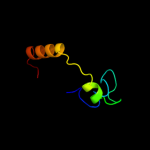
| 10 | c2f42A_
|
|
 |
49.5 |
19 |
PDB header:chaperone
Chain: A: PDB Molecule:stip1 homology and u-box containing protein 1;
PDBTitle: dimerization and u-box domains of zebrafish c-terminal of hsp702 interacting protein
|
| 11 | c3hjhA_
|
|
 |
48.9 |
53 |
PDB header:hydrolase
Chain: A: PDB Molecule:transcription-repair-coupling factor;
PDBTitle: a rigid n-terminal clamp restrains the motor domains of the bacterial2 transcription-repair coupling factor
|
| 12 | d3bz6a2
|
|
 |
32.7 |
30 |
Fold:DNA/RNA-binding 3-helical bundle
Superfamily:"Winged helix" DNA-binding domain
Family:PSPTO2686-like |
| 13 | c3pg6D_
|
|
 |
32.6 |
25 |
PDB header:ligase
Chain: D: PDB Molecule:e3 ubiquitin-protein ligase dtx3l;
PDBTitle: the carboxyl terminal domain of human deltex 3-like
|
| 14 | d2b2na1
|
|
 |
31.1 |
54 |
Fold:P-loop containing nucleoside triphosphate hydrolases
Superfamily:P-loop containing nucleoside triphosphate hydrolases
Family:Tandem AAA-ATPase domain |
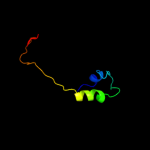
| 15 | d1j6ra_
|
|
 |
31.0 |
27 |
Fold:Methionine synthase activation domain-like
Superfamily:Methionine synthase activation domain-like
Family:Hypothetical protein TM0269 |
| 16 | d2eyqa4
|
|
 |
30.7 |
54 |
Fold:P-loop containing nucleoside triphosphate hydrolases
Superfamily:P-loop containing nucleoside triphosphate hydrolases
Family:Tandem AAA-ATPase domain |
| 17 | d2o4ta1
|
|
 |
29.2 |
13 |
Fold:Left-handed superhelix
Superfamily:BH3980-like
Family:BH3980-like |
| 18 | c3gzfD_
|
|
 |
28.7 |
18 |
PDB header:viral protein
Chain: D: PDB Molecule:replicase polyprotein 1ab;
PDBTitle: structure of the c-terminal domain of nsp4 from feline coronavirus
|
| 19 | c3rv2B_
|
|
 |
27.0 |
18 |
PDB header:transferase
Chain: B: PDB Molecule:s-adenosylmethionine synthase;
PDBTitle: crystal structure of s-adenosylmethionine synthetase from2 mycobacterium marinum
|
| 20 | c3fpnB_
|
|
 |
26.2 |
31 |
PDB header:dna binding protein
Chain: B: PDB Molecule:geobacillus stearothermophilus uvrb interaction
PDBTitle: crystal structure of uvra-uvrb interaction domains
|
| 21 | c3ty1B_ |
|
not modelled |
19.7 |
16 |
PDB header:isomerase
Chain: B: PDB Molecule:hypothetical aldose 1-epimerase;
PDBTitle: crystal structure of a hypothetical aldose 1-epimerase (kpn_04629)2 from klebsiella pneumoniae subsp. pneumoniae mgh 78578 at 1.90 a3 resolution
|
| 22 | d1nb9a_ |
|
not modelled |
16.4 |
10 |
Fold:Reductase/isomerase/elongation factor common domain
Superfamily:Riboflavin kinase-like
Family:ATP-dependent riboflavin kinase-like |
| 23 | d2pvaa_ |
|
not modelled |
15.6 |
18 |
Fold:Ntn hydrolase-like
Superfamily:N-terminal nucleophile aminohydrolases (Ntn hydrolases)
Family:Penicillin V acylase |
| 24 | d1t5la1 |
|
not modelled |
15.5 |
31 |
Fold:P-loop containing nucleoside triphosphate hydrolases
Superfamily:P-loop containing nucleoside triphosphate hydrolases
Family:Tandem AAA-ATPase domain |
| 25 | c2hezB_ |
|
not modelled |
15.3 |
21 |
PDB header:hydrolase
Chain: B: PDB Molecule:bile salt hydrolase;
PDBTitle: bifidobacterium longum bile salt hydrolase
|
| 26 | c3bz6A_ |
|
not modelled |
15.1 |
30 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:upf0502 protein pspto_2686;
PDBTitle: crystal structure of a conserved protein of unknown function from2 pseudomonas syringae pv. tomato str. dc3000
|
| 27 | c3op1A_ |
|
not modelled |
14.5 |
21 |
PDB header:transferase
Chain: A: PDB Molecule:macrolide-efflux protein;
PDBTitle: crystal structure of macrolide-efflux protein sp_1110 from2 streptococcus pneumoniae
|
| 28 | d1n08a_ |
|
not modelled |
13.9 |
10 |
Fold:Reductase/isomerase/elongation factor common domain
Superfamily:Riboflavin kinase-like
Family:ATP-dependent riboflavin kinase-like |
| 29 | c3bnwA_ |
|
not modelled |
12.9 |
16 |
PDB header:transferase
Chain: A: PDB Molecule:riboflavin kinase, putative;
PDBTitle: crystal structure of riboflavin kinase from trypanosoma brucei
|
| 30 | d1tyja1 |
|
not modelled |
12.6 |
25 |
Fold:Common fold of diphtheria toxin/transcription factors/cytochrome f
Superfamily:Carbohydrate-binding domain
Family:Cellulose-binding domain family III |
| 31 | d1zv9a1 |
|
not modelled |
12.4 |
32 |
Fold:Common fold of diphtheria toxin/transcription factors/cytochrome f
Superfamily:Carbohydrate-binding domain
Family:Cellulose-binding domain family III |
| 32 | c3izcW_ |
|
not modelled |
12.2 |
22 |
PDB header:ribosome
Chain: W: PDB Molecule:60s ribosomal protein rpl22 (l22e);
PDBTitle: localization of the large subunit ribosomal proteins into a 6.1 a2 cryo-em map of saccharomyces cerevisiae translating 80s ribosome
|
| 33 | c2eyqA_ |
|
not modelled |
11.8 |
54 |
PDB header:hydrolase
Chain: A: PDB Molecule:transcription-repair coupling factor;
PDBTitle: crystal structure of escherichia coli transcription-repair2 coupling factor
|
| 34 | c2im9A_ |
|
not modelled |
11.2 |
19 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:hypothetical protein;
PDBTitle: crystal structure of protein lpg0564 from legionella pneumophila str.2 philadelphia 1, pfam duf1460
|
| 35 | d2im9a1 |
|
not modelled |
11.2 |
19 |
Fold:Cysteine proteinases
Superfamily:Cysteine proteinases
Family:Lpg0564-like |
| 36 | c3h8mB_ |
|
not modelled |
11.0 |
19 |
PDB header:transferase
Chain: B: PDB Molecule:ephrin type-a receptor 7;
PDBTitle: sam domain of human ephrin type-a receptor 7 (epha7)
|
| 37 | c3lhiA_ |
|
not modelled |
10.5 |
21 |
PDB header:hydrolase
Chain: A: PDB Molecule:putative 6-phosphogluconolactonase;
PDBTitle: crystal structure of putative 6-2 phosphogluconolactonase(yp_207848.1) from neisseria3 gonorrhoeae fa 1090 at 1.33 a resolution
|
| 38 | c4a1dM_ |
|
not modelled |
10.3 |
26 |
PDB header:ribosome
Chain: M: PDB Molecule:ribosomal protein l22;
PDBTitle: t.thermophila 60s ribosomal subunit in complex with initiation2 factor 6. this file contains 26s rrna and proteins of3 molecule 4.
|
| 39 | c3kkaD_ |
|
not modelled |
10.1 |
23 |
PDB header:transferase
Chain: D: PDB Molecule:ephrin type-a receptor 2;
PDBTitle: co-crystal structure of the sam domains of epha1 and epha2
|
| 40 | c2b59A_ |
|
not modelled |
9.7 |
44 |
PDB header:hydrolase/structural protein
Chain: A: PDB Molecule:cog1196: chromosome segregation atpases;
PDBTitle: the type ii cohesin dockerin complex
|
| 41 | d2c2la2 |
|
not modelled |
9.7 |
17 |
Fold:RING/U-box
Superfamily:RING/U-box
Family:U-box |
| 42 | d2bm3a1 |
|
not modelled |
9.6 |
44 |
Fold:Common fold of diphtheria toxin/transcription factors/cytochrome f
Superfamily:Carbohydrate-binding domain
Family:Cellulose-binding domain family III |
| 43 | c2kreA_ |
|
not modelled |
9.6 |
17 |
PDB header:protein binding
Chain: A: PDB Molecule:ubiquitin conjugation factor e4 b;
PDBTitle: solution structure of e4b/ufd2a u-box domain
|
| 44 | c3o2kA_ |
|
not modelled |
9.5 |
23 |
PDB header:transferase
Chain: A: PDB Molecule:brevianamide f prenyltransferase;
PDBTitle: crystal structure of brevianamide f prenyltransferase complexed with2 brevianamide f and dimethylallyl s-thiolodiphosphate
|
| 45 | c2x0kB_ |
|
not modelled |
9.4 |
11 |
PDB header:transferase
Chain: B: PDB Molecule:riboflavin biosynthesis protein ribf;
PDBTitle: crystal structure of modular fad synthetase from2 corynebacterium ammoniagenes
|
| 46 | c1m0kA_ |
|
not modelled |
9.3 |
50 |
PDB header:ion transport
Chain: A: PDB Molecule:bacteriorhodopsin;
PDBTitle: bacteriorhodopsin k intermediate at 1.43 a resolution
|
| 47 | d1m0ka_ |
|
not modelled |
9.3 |
50 |
Fold:Family A G protein-coupled receptor-like
Superfamily:Family A G protein-coupled receptor-like
Family:Bacteriorhodopsin-like |
| 48 | c3fnkA_ |
|
not modelled |
9.2 |
56 |
PDB header:structural protein
Chain: A: PDB Molecule:cellulosomal scaffoldin adaptor protein b;
PDBTitle: crystal structure of the second type ii cohesin module from2 the cellulosomal adaptor scaa scaffoldin of acetivibrio3 cellulolyticus
|
| 49 | c2y9xC_ |
|
not modelled |
9.1 |
14 |
PDB header:oxidoreductase
Chain: C: PDB Molecule:polyphenol oxidase;
PDBTitle: crystal structure of ppo3, a tyrosinase from agaricus bisporus, in2 deoxy-form that contains additional unknown lectin-like subunit,3 with inhibitor tropolone
|
| 50 | c2xfmA_ |
|
not modelled |
8.6 |
33 |
PDB header:rna/protein
Chain: A: PDB Molecule:piwi-like protein 1;
PDBTitle: complex structure of the miwi paz domain bound to methylated single2 stranded rna
|
| 51 | d1gwya_ |
|
not modelled |
8.2 |
12 |
Fold:Cytolysin/lectin
Superfamily:Cytolysin/lectin
Family:Anemone pore-forming cytolysin |
| 52 | d2phcb2 |
|
not modelled |
8.1 |
29 |
Fold:DCoH-like
Superfamily:PH0987 N-terminal domain-like
Family:PH0987 N-terminal domain-like |
| 53 | d1iyca_ |
|
not modelled |
8.0 |
50 |
Fold:Invertebrate chitin-binding proteins
Superfamily:Invertebrate chitin-binding proteins
Family:Antifungal peptide scarabaecin |
| 54 | c3ghpA_ |
|
not modelled |
7.9 |
56 |
PDB header:structural protein
Chain: A: PDB Molecule:cellulosomal scaffoldin adaptor protein b;
PDBTitle: structure of the second type ii cohesin module from the2 adaptor scaa scaffoldin of acetivibrio cellulolyticus3 (including long c-terminal linker)
|
| 55 | d1v2ya_ |
|
not modelled |
7.8 |
25 |
Fold:beta-Grasp (ubiquitin-like)
Superfamily:Ubiquitin-like
Family:Ubiquitin-related |
| 56 | c3i4zA_ |
|
not modelled |
7.6 |
23 |
PDB header:transferase
Chain: A: PDB Molecule:tryptophan dimethylallyltransferase;
PDBTitle: crystal structure of the dimethylallyl tryptophan synthase fgapt2 from2 aspergillus fumigatus
|
| 57 | d2cqpa1 |
|
not modelled |
7.6 |
9 |
Fold:Ferredoxin-like
Superfamily:RNA-binding domain, RBD
Family:Canonical RBD |
| 58 | c1t6zB_ |
|
not modelled |
7.6 |
14 |
PDB header:transferase
Chain: B: PDB Molecule:riboflavin kinase/fmn adenylyltransferase;
PDBTitle: crystal structure of riboflavin bound tm379
|
| 59 | c2quoA_ |
|
not modelled |
7.4 |
29 |
PDB header:toxin
Chain: A: PDB Molecule:heat-labile enterotoxin b chain;
PDBTitle: crystal structure of c terminal fragment of clostridium2 perfringens enterotoxin
|
| 60 | c3htkC_ |
|
not modelled |
7.3 |
17 |
PDB header:recombination/replication/ligase
Chain: C: PDB Molecule:e3 sumo-protein ligase mms21;
PDBTitle: crystal structure of mms21 and smc5 complex
|
| 61 | d1p42a1 |
|
not modelled |
6.9 |
10 |
Fold:Ribosomal protein S5 domain 2-like
Superfamily:Ribosomal protein S5 domain 2-like
Family:UDP-3-O-[3-hydroxymyristoyl] N-acetylglucosamine deacetylase LpxC |
| 62 | c3dclC_ |
|
not modelled |
6.8 |
10 |
PDB header:structural genomics, unknown function
Chain: C: PDB Molecule:tm1086;
PDBTitle: crystal structure of tm1086
|
| 63 | d2gpia1 |
|
not modelled |
6.8 |
27 |
Fold:Shew3726-like
Superfamily:Shew3726-like
Family:Shew3726-like |
| 64 | c2dbhA_ |
|
not modelled |
6.6 |
56 |
PDB header:signaling protein
Chain: A: PDB Molecule:tumor necrosis factor receptor superfamily
PDBTitle: solution structure of the carboxyl-terminal card-like2 domain in human tnfr-related death receptor-6
|
| 65 | d1mxaa2 |
|
not modelled |
6.5 |
33 |
Fold:S-adenosylmethionine synthetase
Superfamily:S-adenosylmethionine synthetase
Family:S-adenosylmethionine synthetase |
| 66 | d2hiya1 |
|
not modelled |
6.4 |
20 |
Fold:SP0830-like
Superfamily:SP0830-like
Family:SP0830-like |
| 67 | c1zc1A_ |
|
not modelled |
6.1 |
21 |
PDB header:protein turnover
Chain: A: PDB Molecule:ubiquitin fusion degradation protein 1;
PDBTitle: ufd1 exhibits the aaa-atpase fold with two distinct2 ubiquitin interaction sites
|
| 68 | d2d9ra1 |
|
not modelled |
5.9 |
25 |
Fold:Double-split beta-barrel
Superfamily:AF2212/PG0164-like
Family:PG0164-like |
| 69 | d1ogya1 |
|
not modelled |
5.8 |
25 |
Fold:Double psi beta-barrel
Superfamily:ADC-like
Family:Formate dehydrogenase/DMSO reductase, C-terminal domain |
| 70 | d1c8sa_ |
|
not modelled |
5.8 |
50 |
Fold:Family A G protein-coupled receptor-like
Superfamily:Family A G protein-coupled receptor-like
Family:Bacteriorhodopsin-like |
| 71 | c3vcbB_ |
|
not modelled |
5.8 |
16 |
PDB header:viral protein
Chain: B: PDB Molecule:rna-directed rna polymerase;
PDBTitle: c425s mutant of the c-terminal cytoplasmic domain of non-structural2 protein 4 from mouse hepatitis virus a59
|
| 72 | d2baya1 |
|
not modelled |
5.8 |
40 |
Fold:RING/U-box
Superfamily:RING/U-box
Family:U-box |
| 73 | d2p02a2 |
|
not modelled |
5.6 |
40 |
Fold:S-adenosylmethionine synthetase
Superfamily:S-adenosylmethionine synthetase
Family:S-adenosylmethionine synthetase |
| 74 | c1zbe1_ |
|
not modelled |
5.4 |
23 |
PDB header:virus
Chain: 1: PDB Molecule:coat protein vp1;
PDBTitle: foot-and mouth disease virus serotype a1061
|
| 75 | d2dira1 |
|
not modelled |
5.4 |
15 |
Fold:THUMP domain
Superfamily:THUMP domain-like
Family:Minimal THUMP |
| 76 | d1x6ma_ |
|
not modelled |
5.3 |
25 |
Fold:Mss4-like
Superfamily:Mss4-like
Family:Glutathione-dependent formaldehyde-activating enzyme, Gfa |
| 77 | d1usua_ |
|
not modelled |
5.2 |
21 |
Fold:Ribosomal protein S5 domain 2-like
Superfamily:Ribosomal protein S5 domain 2-like
Family:Hsp90 middle domain |
| 78 | c2hz7A_ |
|
not modelled |
5.2 |
17 |
PDB header:ligase
Chain: A: PDB Molecule:glutaminyl-trna synthetase;
PDBTitle: crystal structure of the glutaminyl-trna synthetase from2 deinococcus radiodurans
|
| 79 | d1qm4a2 |
|
not modelled |
5.1 |
40 |
Fold:S-adenosylmethionine synthetase
Superfamily:S-adenosylmethionine synthetase
Family:S-adenosylmethionine synthetase |