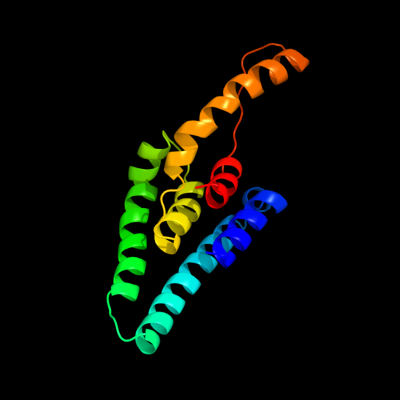
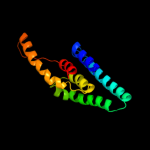
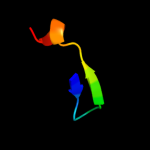
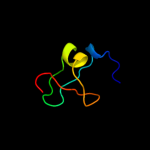
1 c3s0xB_
98.2
17
PDB header: hydrolaseChain: B: PDB Molecule: peptidase a24b, flak domain protein;PDBTitle: the crystal structure of gxgd membrane protease flak
2 c2k5cA_
80.8
6
PDB header: metal binding proteinChain: A: PDB Molecule: uncharacterized protein pf0385;PDBTitle: nmr structure for pf0385
3 c3py7A_
80.8
12
PDB header: viral proteinChain: A: PDB Molecule: maltose-binding periplasmic protein,paxillin ld1,protein e6PDBTitle: crystal structure of full-length bovine papillomavirus oncoprotein e62 in complex with ld1 motif of paxillin at 2.3a resolution
4 c2owoA_
64.3
4
PDB header: ligase/dnaChain: A: PDB Molecule: dna ligase;PDBTitle: last stop on the road to repair: structure of e.coli dna ligase bound2 to nicked dna-adenylate
5 c1v9pB_
63.6
0
PDB header: ligaseChain: B: PDB Molecule: dna ligase;PDBTitle: crystal structure of nad+-dependent dna ligase
6 d1dgsa1
62.3
0
Fold: SAM domain-likeSuperfamily: RuvA domain 2-likeFamily: NAD+-dependent DNA ligase, domain 37 d1pfva3
60.0
7
Fold: Rubredoxin-likeSuperfamily: Methionyl-tRNA synthetase (MetRS), Zn-domainFamily: Methionyl-tRNA synthetase (MetRS), Zn-domain8 c2lcqA_
55.3
7
PDB header: metal binding proteinChain: A: PDB Molecule: putative toxin vapc6;PDBTitle: solution structure of the endonuclease nob1 from p.horikoshii
9 c1dgsB_
54.4
4
PDB header: ligaseChain: B: PDB Molecule: dna ligase;PDBTitle: crystal structure of nad+-dependent dna ligase from t.2 filiformis
10 d1vd4a_
53.9
5
Fold: Rubredoxin-likeSuperfamily: Zinc beta-ribbonFamily: Transcriptional factor domain11 d2fk4a1
53.0
10
Fold: E6 C-terminal domain-likeSuperfamily: E6 C-terminal domain-likeFamily: E6 C-terminal domain-like12 d2jnea1
48.2
14
Fold: Rubredoxin-likeSuperfamily: YfgJ-likeFamily: YfgJ-like13 c2jneA_
48.2
14
PDB header: metal binding proteinChain: A: PDB Molecule: hypothetical protein yfgj;PDBTitle: nmr structure of e.coli yfgj modelled with two zn+2 bound.2 northeast structural genomics consortium target er317.
14 d2fiya1
45.8
13
Fold: FdhE-likeSuperfamily: FdhE-likeFamily: FdhE-like15 d2gnra1
44.2
6
Fold: OB-foldSuperfamily: Nucleic acid-binding proteinsFamily: SSO2064-like16 d2baya1
43.9
6
Fold: RING/U-boxSuperfamily: RING/U-boxFamily: U-box17 c2xeuA_
43.5
15
PDB header: transcriptionChain: A: PDB Molecule: ring finger protein 4;PDBTitle: ring domain
18 d1faqa_
41.0
21
Fold: Cysteine-rich domainSuperfamily: Cysteine-rich domainFamily: Protein kinase cysteine-rich domain (cys2, phorbol-binding domain)19 c3pfqA_
38.2
7
PDB header: transferaseChain: A: PDB Molecule: protein kinase c beta type;PDBTitle: crystal structure and allosteric activation of protein kinase c beta2 ii
20 d1rmda2
37.5
8
Fold: RING/U-boxSuperfamily: RING/U-boxFamily: RING finger domain, C3HC421 c2kr6A_
not modelled
36.9
17
PDB header: hydrolaseChain: A: PDB Molecule: presenilin-1;PDBTitle: solution structure of presenilin-1 ctf subunit
22 d2ctda2
not modelled
36.0
22
Fold: beta-beta-alpha zinc fingersSuperfamily: beta-beta-alpha zinc fingersFamily: Classic zinc finger, C2H223 c3lrqB_
not modelled
35.6
5
PDB header: ligaseChain: B: PDB Molecule: e3 ubiquitin-protein ligase trim37;PDBTitle: crystal structure of the u-box domain of human ubiquitin-2 protein ligase (e3), northeast structural genomics3 consortium target hr4604d.
24 c2f42A_
not modelled
34.6
5
PDB header: chaperoneChain: A: PDB Molecule: stip1 homology and u-box containing protein 1;PDBTitle: dimerization and u-box domains of zebrafish c-terminal of hsp702 interacting protein
25 c2jmdA_
not modelled
33.1
5
PDB header: ligaseChain: A: PDB Molecule: tnf receptor-associated factor 6;PDBTitle: solution structure of the ring domain of human traf6
26 d1iyca_
not modelled
31.5
21
Fold: Invertebrate chitin-binding proteinsSuperfamily: Invertebrate chitin-binding proteinsFamily: Antifungal peptide scarabaecin27 c3cngC_
not modelled
31.3
0
PDB header: hydrolaseChain: C: PDB Molecule: nudix hydrolase;PDBTitle: crystal structure of nudix hydrolase from nitrosomonas europaea
28 d2cu8a1
not modelled
28.8
10
Fold: Glucocorticoid receptor-like (DNA-binding domain)Superfamily: Glucocorticoid receptor-like (DNA-binding domain)Family: LIM domain29 c3ky9B_
not modelled
28.8
4
PDB header: apoptosisChain: B: PDB Molecule: proto-oncogene vav;PDBTitle: autoinhibited vav1
30 c2eciA_
not modelled
28.7
5
PDB header: metal binding proteinChain: A: PDB Molecule: tnf receptor-associated factor 6;PDBTitle: solution structure of the ring domain of the human tnf2 receptor-associated factor 6 protein
31 d1vfya_
not modelled
28.5
19
Fold: FYVE/PHD zinc fingerSuperfamily: FYVE/PHD zinc fingerFamily: FYVE, a phosphatidylinositol-3-phosphate binding domain32 d1kbea_
not modelled
28.3
17
Fold: Cysteine-rich domainSuperfamily: Cysteine-rich domainFamily: Protein kinase cysteine-rich domain (cys2, phorbol-binding domain)33 c2db6A_
not modelled
28.2
3
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: sh3 and cysteine rich domain 3;PDBTitle: solution structure of rsgi ruh-051, a c1 domain of stac32 from human cdna
34 d1dvpa2
not modelled
28.2
9
Fold: FYVE/PHD zinc fingerSuperfamily: FYVE/PHD zinc fingerFamily: FYVE, a phosphatidylinositol-3-phosphate binding domain35 d1s5pa_
not modelled
28.2
16
Fold: DHS-like NAD/FAD-binding domainSuperfamily: DHS-like NAD/FAD-binding domainFamily: Sir2 family of transcriptional regulators36 c2jrpA_
not modelled
28.2
0
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: putative cytoplasmic protein;PDBTitle: solution nmr structure of yfgj from salmonella typhimurium2 modeled with two zn+2 bound, northeast structural genomics3 consortium target str86
37 c3llkA_
not modelled
28.0
10
PDB header: oxidoreductaseChain: A: PDB Molecule: sulfhydryl oxidase 1;PDBTitle: sulfhydryl oxidase fragment of human qsox1
38 d2csha2
not modelled
27.9
18
Fold: beta-beta-alpha zinc fingersSuperfamily: beta-beta-alpha zinc fingersFamily: Classic zinc finger, C2H239 c2klxA_
not modelled
27.4
12
PDB header: oxidoreductaseChain: A: PDB Molecule: glutaredoxin;PDBTitle: solution structure of glutaredoxin from bartonella henselae str.2 houston
40 d1jm7a_
not modelled
26.2
0
Fold: RING/U-boxSuperfamily: RING/U-boxFamily: RING finger domain, C3HC441 c2i5oA_
not modelled
25.9
8
PDB header: transferaseChain: A: PDB Molecule: dna polymerase eta;PDBTitle: solution structure of the ubiquitin-binding zinc finger2 (ubz) domain of the human dna y-polymerase eta
42 d2cona1
not modelled
25.5
18
Fold: Rubredoxin-likeSuperfamily: NOB1 zinc finger-likeFamily: NOB1 zinc finger-like43 d2ct1a1
not modelled
25.1
10
Fold: beta-beta-alpha zinc fingersSuperfamily: beta-beta-alpha zinc fingersFamily: Classic zinc finger, C2H244 d1dsva_
not modelled
25.0
29
Fold: Retrovirus zinc finger-like domainsSuperfamily: Retrovirus zinc finger-like domainsFamily: Retrovirus zinc finger-like domains45 c3zyqA_
not modelled
24.5
7
PDB header: signalingChain: A: PDB Molecule: hepatocyte growth factor-regulated tyrosine kinasePDBTitle: crystal structure of the tandem vhs and fyve domains of hepatocyte2 growth factor-regulated tyrosine kinase substrate (hgs-hrs) at 1.483 a resolution
46 d2c2la2
not modelled
24.1
5
Fold: RING/U-boxSuperfamily: RING/U-boxFamily: U-box47 c3odeA_
not modelled
23.4
7
PDB header: dna binding protein/dnaChain: A: PDB Molecule: poly [adp-ribose] polymerase 1;PDBTitle: human parp-1 zinc finger 2 (zn2) bound to dna
48 d1ptqa_
not modelled
23.3
3
Fold: Cysteine-rich domainSuperfamily: Cysteine-rich domainFamily: Protein kinase cysteine-rich domain (cys2, phorbol-binding domain)49 d2vrda1
not modelled
22.3
0
Fold: beta-beta-alpha zinc fingersSuperfamily: beta-beta-alpha zinc fingersFamily: HkH motif-containing C2H2 finger50 c1dvpA_
not modelled
21.9
7
PDB header: transferaseChain: A: PDB Molecule: hepatocyte growth factor-regulated tyrosinePDBTitle: crystal structure of the vhs and fyve tandem domains of hrs,2 a protein involved in membrane trafficking and signal3 transduction
51 c1rfhA_
not modelled
21.1
22
PDB header: metal binding proteinChain: A: PDB Molecule: ras association (ralgds/af-6) domain family 5;PDBTitle: solution structure of the c1 domain of nore1, a novel ras2 effector
52 d2jnya1
not modelled
21.0
0
Fold: Trm112p-likeSuperfamily: Trm112p-likeFamily: Trm112p-like53 d1x4ka1
not modelled
20.9
20
Fold: Glucocorticoid receptor-like (DNA-binding domain)Superfamily: Glucocorticoid receptor-like (DNA-binding domain)Family: LIM domain54 d1r3jc_
not modelled
20.7
21
Fold: Voltage-gated potassium channelsSuperfamily: Voltage-gated potassium channelsFamily: Voltage-gated potassium channels55 d1wfka_
not modelled
20.5
8
Fold: FYVE/PHD zinc fingerSuperfamily: FYVE/PHD zinc fingerFamily: FYVE, a phosphatidylinositol-3-phosphate binding domain56 d1yfua1
not modelled
20.4
16
Fold: Double-stranded beta-helixSuperfamily: RmlC-like cupinsFamily: 3-hydroxyanthranilic acid dioxygenase-like57 d1tbna_
not modelled
20.2
10
Fold: Cysteine-rich domainSuperfamily: Cysteine-rich domainFamily: Protein kinase cysteine-rich domain (cys2, phorbol-binding domain)58 d2pk7a1
not modelled
20.1
0
Fold: Trm112p-likeSuperfamily: Trm112p-likeFamily: Trm112p-like59 c3ir9A_
not modelled
20.1
3
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: peptide chain release factor subunit 1;PDBTitle: c-terminal domain of peptide chain release factor from2 methanosarcina mazei.
60 c2j6aA_
not modelled
19.9
13
PDB header: transferaseChain: A: PDB Molecule: protein trm112;PDBTitle: crystal structure of s. cerevisiae ynr046w, a zinc finger2 protein from the erf1 methyltransferase complex.
61 d2c2vv1
not modelled
19.8
5
Fold: RING/U-boxSuperfamily: RING/U-boxFamily: U-box62 c2y43B_
not modelled
19.7
5
PDB header: ligaseChain: B: PDB Molecule: e3 ubiquitin-protein ligase rad18;PDBTitle: rad18 ubiquitin ligase ring domain structure
63 d2hf1a1
not modelled
19.5
0
Fold: Trm112p-likeSuperfamily: Trm112p-likeFamily: Trm112p-like64 d2gmga1
not modelled
19.1
0
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: PF0610-like65 c3nznA_
not modelled
19.1
10
PDB header: oxidoreductaseChain: A: PDB Molecule: glutaredoxin;PDBTitle: the crystal structure of the glutaredoxin from methanosarcina mazei2 go1
66 c2ecyA_
not modelled
18.9
6
PDB header: apoptosisChain: A: PDB Molecule: tnf receptor-associated factor 3;PDBTitle: solution structure of the zinc finger, c3hc4 type (ring2 finger)" domain of tnf receptor-associated factor 3
67 c2egpA_
not modelled
18.7
4
PDB header: antiviral proteinChain: A: PDB Molecule: tripartite motif-containing protein 34;PDBTitle: solution structure of the ring-finger domain from human2 tripartite motif protein 34
68 d2yrka1
not modelled
18.5
15
Fold: beta-beta-alpha zinc fingersSuperfamily: beta-beta-alpha zinc fingersFamily: HkH motif-containing C2H2 finger69 c2gb5B_
not modelled
18.5
0
PDB header: hydrolaseChain: B: PDB Molecule: nadh pyrophosphatase;PDBTitle: crystal structure of nadh pyrophosphatase (ec 3.6.1.22) (1790429) from2 escherichia coli k12 at 2.30 a resolution
70 c2dmjA_
not modelled
18.4
0
PDB header: transferaseChain: A: PDB Molecule: poly (adp-ribose) polymerase family, member 1;PDBTitle: solution structure of the first zf-parp domain of human2 poly(adp-ribose)polymerase-1
71 c2kreA_
not modelled
18.2
11
PDB header: protein bindingChain: A: PDB Molecule: ubiquitin conjugation factor e4 b;PDBTitle: solution structure of e4b/ufd2a u-box domain
72 d1iyma_
not modelled
17.7
0
Fold: RING/U-boxSuperfamily: RING/U-boxFamily: RING finger domain, C3HC473 c2jvyA_
not modelled
17.6
19
PDB header: metal binding proteinChain: A: PDB Molecule: nf-kappa-b essential modulator;PDBTitle: solution structure of the eda-id-related c417f mutant of2 human nemo zinc finger
74 c3glsC_
not modelled
17.6
0
PDB header: hydrolaseChain: C: PDB Molecule: nad-dependent deacetylase sirtuin-3,PDBTitle: crystal structure of human sirt3
75 c1z2qA_
not modelled
17.1
14
PDB header: membrane proteinChain: A: PDB Molecule: lm5-1;PDBTitle: high-resolution solution structure of the lm5-1 fyve domain2 from leishmania major
76 d2dara2
not modelled
16.9
5
Fold: Glucocorticoid receptor-like (DNA-binding domain)Superfamily: Glucocorticoid receptor-like (DNA-binding domain)Family: LIM domain77 d1tdza3
not modelled
16.9
0
Fold: Glucocorticoid receptor-like (DNA-binding domain)Superfamily: Glucocorticoid receptor-like (DNA-binding domain)Family: C-terminal, Zn-finger domain of MutM-like DNA repair proteins78 d1t1ha_
not modelled
16.7
11
Fold: RING/U-boxSuperfamily: RING/U-boxFamily: U-box79 c1jocA_
not modelled
16.7
14
PDB header: membrane proteinChain: A: PDB Molecule: early endosomal autoantigen 1;PDBTitle: eea1 homodimer of c-terminal fyve domain bound to inositol2 1,3-diphosphate
80 d1rutx3
not modelled
16.7
7
Fold: Glucocorticoid receptor-like (DNA-binding domain)Superfamily: Glucocorticoid receptor-like (DNA-binding domain)Family: LIM domain81 c3mhsE_
not modelled
16.6
21
PDB header: hydrolase/transcription regulator/proteiChain: E: PDB Molecule: saga-associated factor 73;PDBTitle: structure of the saga ubp8/sgf11/sus1/sgf73 dub module bound to2 ubiquitin aldehyde
82 d1r2za3
not modelled
16.4
0
Fold: Glucocorticoid receptor-like (DNA-binding domain)Superfamily: Glucocorticoid receptor-like (DNA-binding domain)Family: C-terminal, Zn-finger domain of MutM-like DNA repair proteins83 c2jr6A_
not modelled
16.4
0
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: upf0434 protein nma0874;PDBTitle: solution structure of upf0434 protein nma0874. northeast structural2 genomics target mr32
84 c3htkC_
not modelled
16.3
13
PDB header: recombination/replication/ligaseChain: C: PDB Molecule: e3 sumo-protein ligase mms21;PDBTitle: crystal structure of mms21 and smc5 complex
85 d1xx6a2
not modelled
16.0
22
Fold: Glucocorticoid receptor-like (DNA-binding domain)Superfamily: Glucocorticoid receptor-like (DNA-binding domain)Family: Type II thymidine kinase zinc finger86 d1vqo11
not modelled
16.0
11
Fold: Rubredoxin-likeSuperfamily: Zn-binding ribosomal proteinsFamily: Ribosomal protein L37e87 c2e7pC_
not modelled
15.9
4
PDB header: electron transportChain: C: PDB Molecule: glutaredoxin;PDBTitle: crystal structure of the holo form of glutaredoxin c1 from populus2 tremula x tremuloides
88 c2e73A_
not modelled
15.7
7
PDB header: transferaseChain: A: PDB Molecule: protein kinase c gamma type;PDBTitle: solution structure of the phorbol esters/diacylglycerol2 binding domain of protein kinase c gamma
89 d1lkoa2
not modelled
15.4
11
Fold: Rubredoxin-likeSuperfamily: Rubredoxin-likeFamily: Rubredoxin90 c2zaeB_
not modelled
15.3
4
PDB header: hydrolaseChain: B: PDB Molecule: ribonuclease p protein component 4;PDBTitle: crystal structure of protein ph1601p in complex with protein ph1771p2 of archaeal ribonuclease p from pyrococcus horikoshii ot3
91 c3gkxB_
not modelled
15.3
17
PDB header: structural genomics, unknown functionChain: B: PDB Molecule: putative arsc family related protein;PDBTitle: crystal structure of putative arsc family related protein from2 bacteroides fragilis
92 c2js4A_
not modelled
15.2
0
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: upf0434 protein bb2007;PDBTitle: solution nmr structure of bordetella bronchiseptica protein2 bb2007. northeast structural genomics consortium target3 bor54
93 c2h0dB_
not modelled
15.2
16
PDB header: metal binding protein/ligaseChain: B: PDB Molecule: ubiquitin ligase protein ring2;PDBTitle: structure of a bmi-1-ring1b polycomb group ubiquitin ligase complex
94 c3nw0A_
not modelled
15.0
0
PDB header: metal binding proteinChain: A: PDB Molecule: non-structural maintenance of chromosomes element 1PDBTitle: crystal structure of mageg1 and nse1 complex
95 c3i2dA_
not modelled
15.0
13
PDB header: ligaseChain: A: PDB Molecule: e3 sumo-protein ligase siz1;PDBTitle: crystal structure of s. cerevisiae sumo e3 ligase siz1
96 d1yc5a1
not modelled
15.0
9
Fold: DHS-like NAD/FAD-binding domainSuperfamily: DHS-like NAD/FAD-binding domainFamily: Sir2 family of transcriptional regulators97 d1imla2
not modelled
14.8
9
Fold: Glucocorticoid receptor-like (DNA-binding domain)Superfamily: Glucocorticoid receptor-like (DNA-binding domain)Family: LIM domain98 c1v9xA_
not modelled
14.7
0
PDB header: transferaseChain: A: PDB Molecule: poly (adp-ribose) polymerase;PDBTitle: solution structure of the first zn-finger domain of2 poly(adp-ribose) polymerase-1
99 d1q1aa_
not modelled
14.7
0
Fold: DHS-like NAD/FAD-binding domainSuperfamily: DHS-like NAD/FAD-binding domainFamily: Sir2 family of transcriptional regulators