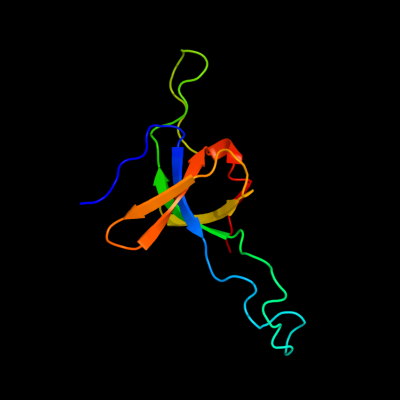
| 1 | d1aono_
|
|
 |
100.0 |
100 |
Fold:GroES-like
Superfamily:GroES-like
Family:GroES |
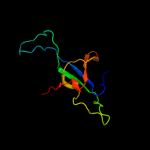
| 2 | d1p3ha_
|
|
 |
100.0 |
45 |
Fold:GroES-like
Superfamily:GroES-like
Family:GroES |
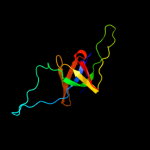
| 3 | d1we3o_
|
|
 |
100.0 |
53 |
Fold:GroES-like
Superfamily:GroES-like
Family:GroES |
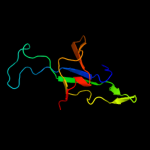
| 4 | d1lepa_
|
|
 |
99.9 |
48 |
Fold:GroES-like
Superfamily:GroES-like
Family:GroES |
| 5 | c3nx6A_
|
|
 |
99.9 |
49 |
PDB header:chaperone
Chain: A: PDB Molecule:10kda chaperonin;
PDBTitle: crystal structure of co-chaperonin, groes (xoo4289) from xanthomonas2 oryzae pv. oryzae kacc10331
|
| 6 | d1g31a_
|
|
 |
95.7 |
19 |
Fold:GroES-like
Superfamily:GroES-like
Family:GroES |
| 7 | c1p82A_
|
|
 |
76.7 |
43 |
PDB header:chaperone
Chain: A: PDB Molecule:10 kda chaperonin;
PDBTitle: nmr structure of 1-25 fragment of mycobacterium2 tuberculosis cpn10
|
| 8 | c1yqxB_
|
|
 |
76.0 |
50 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:sinapyl alcohol dehydrogenase;
PDBTitle: sinapyl alcohol dehydrogenase at 2.5 angstrom resolution
|
| 9 | c1lluD_
|
|
 |
72.3 |
50 |
PDB header:oxidoreductase
Chain: D: PDB Molecule:alcohol dehydrogenase;
PDBTitle: the ternary complex of pseudomonas aeruginosa alcohol2 dehydrogenase with its coenzyme and weak substrate
|
| 10 | c4a10A_
|
|
 |
71.3 |
45 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:octenoyl-coa reductase/carboxylase;
PDBTitle: apo-structure of 2-octenoyl-coa carboxylase reductase cinf from2 streptomyces sp.
|
| 11 | c2ejvA_
|
|
 |
70.3 |
36 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:l-threonine 3-dehydrogenase;
PDBTitle: crystal structure of threonine 3-dehydrogenase complexed with nad+
|
| 12 | c1kevB_
|
|
 |
70.2 |
39 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:nadp-dependent alcohol dehydrogenase;
PDBTitle: structure of nadp-dependent alcohol dehydrogenase
|
| 13 | c1rjwA_
|
|
 |
69.9 |
48 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:alcohol dehydrogenase;
PDBTitle: crystal structure of nad(+)-dependent alcohol dehydrogenase2 from bacillus stearothermophilus strain lld-r
|
| 14 | c3m6iA_
|
|
 |
69.6 |
45 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:l-arabinitol 4-dehydrogenase;
PDBTitle: l-arabinitol 4-dehydrogenase
|
| 15 | c1kolA_
|
|
 |
69.0 |
38 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:formaldehyde dehydrogenase;
PDBTitle: crystal structure of formaldehyde dehydrogenase
|
| 16 | c2ouiB_
|
|
 |
68.9 |
32 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:nadp-dependent alcohol dehydrogenase;
PDBTitle: d275p mutant of alcohol dehydrogenase from protozoa entamoeba2 histolytica
|
| 17 | c1piwA_
|
|
 |
68.4 |
35 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:hypothetical zinc-type alcohol dehydrogenase-
PDBTitle: apo and holo structures of an nadp(h)-dependent cinnamyl2 alcohol dehydrogenase from saccharomyces cerevisiae
|
| 18 | d1uufa1
|
|
 |
67.7 |
35 |
Fold:GroES-like
Superfamily:GroES-like
Family:Alcohol dehydrogenase-like, N-terminal domain |
| 19 | c1pl6A_
|
|
 |
67.7 |
19 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:sorbitol dehydrogenase;
PDBTitle: human sdh/nadh/inhibitor complex
|
| 20 | c2hcyD_
|
|
 |
67.2 |
30 |
PDB header:oxidoreductase
Chain: D: PDB Molecule:alcohol dehydrogenase 1;
PDBTitle: yeast alcohol dehydrogenase i, saccharomyces cerevisiae fermentative2 enzyme
|
| 21 | c1r37B_ |
|
not modelled |
66.7 |
33 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:nad-dependent alcohol dehydrogenase;
PDBTitle: alcohol dehydrogenase from sulfolobus solfataricus2 complexed with nad(h) and 2-ethoxyethanol
|
| 22 | c1h2bA_ |
|
not modelled |
66.1 |
30 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:alcohol dehydrogenase;
PDBTitle: crystal structure of the alcohol dehydrogenase from the2 hyperthermophilic archaeon aeropyrum pernix at 1.65a3 resolution
|
| 23 | c2cf5A_ |
|
not modelled |
65.9 |
44 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:cinnamyl alcohol dehydrogenase;
PDBTitle: crystal structures of the arabidopsis cinnamyl alcohol2 dehydrogenases, atcad5
|
| 24 | c2xaaC_ |
|
not modelled |
65.0 |
36 |
PDB header:oxidoreductase
Chain: C: PDB Molecule:secondary alcohol dehydrogenase;
PDBTitle: alcohol dehydrogenase adh-'a' from rhodococcus ruber dsm2 44541 at ph 8.5 in complex with nad and butane-1,4-diol
|
| 25 | c3krtC_ |
|
not modelled |
63.8 |
30 |
PDB header:oxidoreductase
Chain: C: PDB Molecule:crotonyl coa reductase;
PDBTitle: crystal structure of putative crotonyl coa reductase from streptomyces2 coelicolor a3(2)
|
| 26 | c1f8fA_ |
|
not modelled |
63.7 |
44 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:benzyl alcohol dehydrogenase;
PDBTitle: crystal structure of benzyl alcohol dehydrogenase from acinetobacter2 calcoaceticus
|
| 27 | c2eihA_ |
|
not modelled |
63.1 |
48 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:alcohol dehydrogenase;
PDBTitle: crystal structure of nad-dependent alcohol dehydrogenase
|
| 28 | c1e3jA_ |
|
not modelled |
62.2 |
32 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:nadp(h)-dependent ketose reductase;
PDBTitle: ketose reductase (sorbitol dehydrogenase) from silverleaf2 whitefly
|
| 29 | c1vj0B_ |
|
not modelled |
61.4 |
33 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:alcohol dehydrogenase, zinc-containing;
PDBTitle: crystal structure of alcohol dehydrogenase (tm0436) from thermotoga2 maritima at 2.00 a resolution
|
| 30 | c1cdoB_ |
|
not modelled |
60.0 |
36 |
PDB header:oxidoreductase (ch-oh(d)-nad(a))
Chain: B: PDB Molecule:alcohol dehydrogenase;
PDBTitle: alcohol dehydrogenase (e.c.1.1.1.1) (ee isozyme) complexed with2 nicotinamide adenine dinucleotide (nad), and zinc
|
| 31 | c2dphA_ |
|
not modelled |
59.9 |
33 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:formaldehyde dismutase;
PDBTitle: crystal structure of formaldehyde dismutase
|
| 32 | c1ma0B_ |
|
not modelled |
58.7 |
41 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:glutathione-dependent formaldehyde dehydrogenase;
PDBTitle: ternary complex of human glutathione-dependent formaldehyde2 dehydrogenase with nad+ and dodecanoic acid
|
| 33 | c3uogB_ |
|
not modelled |
58.5 |
37 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:alcohol dehydrogenase;
PDBTitle: crystal structure of putative alcohol dehydrogenase from sinorhizobium2 meliloti 1021
|
| 34 | d1pl8a1 |
|
not modelled |
56.9 |
20 |
Fold:GroES-like
Superfamily:GroES-like
Family:Alcohol dehydrogenase-like, N-terminal domain |
| 35 | c2dfvB_ |
|
not modelled |
55.3 |
45 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:probable l-threonine 3-dehydrogenase;
PDBTitle: hyperthermophilic threonine dehydrogenase from pyrococcus horikoshii
|
| 36 | d2eyqa1 |
|
not modelled |
54.9 |
15 |
Fold:SH3-like barrel
Superfamily:CarD-like
Family:CarD-like |
| 37 | c1uufA_ |
|
not modelled |
54.7 |
32 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:zinc-type alcohol dehydrogenase-like protein
PDBTitle: crystal structure of a zinc-type alcohol dehydrogenase-like2 protein yahk
|
| 38 | c3cosD_ |
|
not modelled |
53.8 |
39 |
PDB header:oxidoreductase
Chain: D: PDB Molecule:alcohol dehydrogenase 4;
PDBTitle: crystal structure of human class ii alcohol dehydrogenase (adh4) in2 complex with nad and zn
|
| 39 | c1p0fA_ |
|
not modelled |
51.7 |
36 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:nadp-dependent alcohol dehydrogenase;
PDBTitle: crystal structure of the binary complex: nadp(h)-dependent vertebrate2 alcohol dehydrogenase (adh8) with the cofactor nadp
|
| 40 | d1piwa1 |
|
not modelled |
51.7 |
33 |
Fold:GroES-like
Superfamily:GroES-like
Family:Alcohol dehydrogenase-like, N-terminal domain |
| 41 | c1hf3A_ |
|
not modelled |
51.3 |
45 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:alcohol dehydrogenase e chain;
PDBTitle: atomic x-ray structure of liver alcohol dehydrogenase2 containing cadmium and a hydroxide adduct to nadh
|
| 42 | c3b70A_ |
|
not modelled |
46.6 |
21 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:enoyl reductase;
PDBTitle: crystal structure of aspergillus terreus trans-acting lovastatin2 polyketide enoyl reductase (lovc) with bound nadp
|
| 43 | d2vnud3 |
|
not modelled |
44.2 |
24 |
Fold:OB-fold
Superfamily:Nucleic acid-binding proteins
Family:Cold shock DNA-binding domain-like |
| 44 | c2cdaA_ |
|
not modelled |
41.6 |
33 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:glucose dehydrogenase;
PDBTitle: sulfolobus solfataricus glucose dehydrogenase 1 in complex2 with nadp
|
| 45 | c2h6eA_ |
|
not modelled |
40.8 |
38 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:d-arabinose 1-dehydrogenase;
PDBTitle: crystal structure of the d-arabinose dehydrogenase from sulfolobus2 solfataricus
|
| 46 | d2z1ca1 |
|
not modelled |
40.6 |
37 |
Fold:OB-fold
Superfamily:HupF/HypC-like
Family:HupF/HypC-like |
| 47 | d1kola1 |
|
not modelled |
39.6 |
38 |
Fold:GroES-like
Superfamily:GroES-like
Family:Alcohol dehydrogenase-like, N-terminal domain |
| 48 | d1p0fa1 |
|
not modelled |
37.4 |
21 |
Fold:GroES-like
Superfamily:GroES-like
Family:Alcohol dehydrogenase-like, N-terminal domain |
| 49 | d1x82a_ |
|
not modelled |
36.2 |
16 |
Fold:Double-stranded beta-helix
Superfamily:RmlC-like cupins
Family:Glucose-6-phosphate isomerase, GPI |
| 50 | c3ip1C_ |
|
not modelled |
30.4 |
19 |
PDB header:oxidoreductase
Chain: C: PDB Molecule:alcohol dehydrogenase, zinc-containing;
PDBTitle: structure of putative alcohol dehydrogenase (tm_042) from thermotoga2 maritima
|
| 51 | c2fg0B_ |
|
not modelled |
29.7 |
26 |
PDB header:hydrolase
Chain: B: PDB Molecule:cog0791: cell wall-associated hydrolases (invasion-
PDBTitle: crystal structure of a putative gamma-d-glutamyl-l-diamino acid2 endopeptidase (npun_r0659) from nostoc punctiforme pcc 73102 at 1.793 a resolution
|
| 52 | d1iuza_ |
|
not modelled |
29.0 |
14 |
Fold:Cupredoxin-like
Superfamily:Cupredoxins
Family:Plastocyanin/azurin-like |
| 53 | d1j3pa_ |
|
not modelled |
28.4 |
13 |
Fold:Double-stranded beta-helix
Superfamily:RmlC-like cupins
Family:Glucose-6-phosphate isomerase, GPI |
| 54 | c2j8zA_ |
|
not modelled |
26.3 |
30 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:quinone oxidoreductase;
PDBTitle: crystal structure of human p53 inducible oxidoreductase (2 tp53i3,pig3)
|
| 55 | c2vnuD_ |
|
not modelled |
26.1 |
27 |
PDB header:hydrolase/rna
Chain: D: PDB Molecule:exosome complex exonuclease rrp44;
PDBTitle: crystal structure of sc rrp44
|
| 56 | c1n9gF_ |
|
not modelled |
25.4 |
30 |
PDB header:hydrolase
Chain: F: PDB Molecule:2,4-dienoyl-coa reductase;
PDBTitle: mitochondrial 2-enoyl thioester reductase etr1p/etr2p2 heterodimer from candida tropicalis
|
| 57 | c3n2bD_ |
|
not modelled |
23.4 |
19 |
PDB header:lyase
Chain: D: PDB Molecule:diaminopimelate decarboxylase;
PDBTitle: 1.8 angstrom resolution crystal structure of diaminopimelate2 decarboxylase (lysa) from vibrio cholerae.
|
| 58 | d1uika1 |
|
not modelled |
23.2 |
21 |
Fold:Double-stranded beta-helix
Superfamily:RmlC-like cupins
Family:Germin/Seed storage 7S protein |
| 59 | d1dgwa_ |
|
not modelled |
22.7 |
16 |
Fold:Double-stranded beta-helix
Superfamily:RmlC-like cupins
Family:Germin/Seed storage 7S protein |
| 60 | d2ot2a1 |
|
not modelled |
21.6 |
24 |
Fold:OB-fold
Superfamily:HupF/HypC-like
Family:HupF/HypC-like |
| 61 | c2c0cB_ |
|
not modelled |
21.1 |
29 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:zinc binding alcohol dehydrogenase, domain
PDBTitle: structure of the mgc45594 gene product
|
| 62 | c3npfB_ |
|
not modelled |
20.2 |
31 |
PDB header:hydrolase
Chain: B: PDB Molecule:putative dipeptidyl-peptidase vi;
PDBTitle: crystal structure of a putative dipeptidyl-peptidase vi (bacova_00612)2 from bacteroides ovatus at 1.72 a resolution
|
| 63 | d1jt8a_ |
|
not modelled |
17.7 |
40 |
Fold:OB-fold
Superfamily:Nucleic acid-binding proteins
Family:Cold shock DNA-binding domain-like |
| 64 | c3h41A_ |
|
not modelled |
16.8 |
23 |
PDB header:hydrolase
Chain: A: PDB Molecule:nlp/p60 family protein;
PDBTitle: crystal structure of a nlpc/p60 family protein (bce_2878) from2 bacillus cereus atcc 10987 at 1.79 a resolution
|
| 65 | d2d6fa1 |
|
not modelled |
16.7 |
30 |
Fold:Sm-like fold
Superfamily:GatD N-terminal domain-like
Family:GatD N-terminal domain-like |
| 66 | c2l55A_ |
|
not modelled |
16.3 |
26 |
PDB header:metal binding protein
Chain: A: PDB Molecule:silb,silver efflux protein, mfp component of the three
PDBTitle: solution structure of the c-terminal domain of silb from cupriavidus2 metallidurans
|
| 67 | d1knwa1 |
|
not modelled |
15.1 |
23 |
Fold:Domain of alpha and beta subunits of F1 ATP synthase-like
Superfamily:Alanine racemase C-terminal domain-like
Family:Eukaryotic ODC-like |
| 68 | c2kijA_ |
|
not modelled |
15.0 |
22 |
PDB header:hydrolase
Chain: A: PDB Molecule:copper-transporting atpase 1;
PDBTitle: solution structure of the actuator domain of the copper-2 transporting atpase atp7a
|
| 69 | c3gt2A_ |
|
not modelled |
14.4 |
15 |
PDB header:unknown function
Chain: A: PDB Molecule:putative uncharacterized protein;
PDBTitle: crystal structure of the p60 domain from m. avium2 paratuberculosis antigen map1272c
|
| 70 | c3d3rA_ |
|
not modelled |
14.2 |
20 |
PDB header:chaperone
Chain: A: PDB Molecule:hydrogenase assembly chaperone hypc/hupf;
PDBTitle: crystal structure of the hydrogenase assembly chaperone hypc/hupf2 family protein from shewanella oneidensis mr-1
|
| 71 | c3gazA_ |
|
not modelled |
13.8 |
21 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:alcohol dehydrogenase superfamily protein;
PDBTitle: crystal structure of an alcohol dehydrogenase superfamily protein from2 novosphingobium aromaticivorans
|
| 72 | d2oyza1 |
|
not modelled |
13.8 |
19 |
Fold:Double-stranded beta-helix
Superfamily:RmlC-like cupins
Family:VPA0057-like |
| 73 | d1gppa_ |
|
not modelled |
13.5 |
23 |
Fold:Hedgehog/intein (Hint) domain
Superfamily:Hedgehog/intein (Hint) domain
Family:Intein (protein splicing domain) |
| 74 | d1g7sa1 |
|
not modelled |
13.5 |
16 |
Fold:Reductase/isomerase/elongation factor common domain
Superfamily:Translation proteins
Family:Elongation factors |
| 75 | d1xa0a1 |
|
not modelled |
13.3 |
17 |
Fold:GroES-like
Superfamily:GroES-like
Family:Alcohol dehydrogenase-like, N-terminal domain |
| 76 | c3gjbA_ |
|
not modelled |
13.3 |
21 |
PDB header:biosynthetic protein
Chain: A: PDB Molecule:cytc3;
PDBTitle: cytc3 with fe(ii) and alpha-ketoglutarate
|
| 77 | d1oe1a2 |
|
not modelled |
12.8 |
42 |
Fold:Cupredoxin-like
Superfamily:Cupredoxins
Family:Multidomain cupredoxins |
| 78 | d1mzya2 |
|
not modelled |
12.8 |
58 |
Fold:Cupredoxin-like
Superfamily:Cupredoxins
Family:Multidomain cupredoxins |
| 79 | c2xivA_ |
|
not modelled |
12.6 |
12 |
PDB header:structural protein
Chain: A: PDB Molecule:hypothetical invasion protein;
PDBTitle: structure of rv1477, hypothetical invasion protein of2 mycobacterium tuberculosis
|
| 80 | d1zcea1 |
|
not modelled |
12.6 |
40 |
Fold:PUA domain-like
Superfamily:PUA domain-like
Family:Atu2648/PH1033-like |
| 81 | c1dgsB_ |
|
not modelled |
12.6 |
30 |
PDB header:ligase
Chain: B: PDB Molecule:dna ligase;
PDBTitle: crystal structure of nad+-dependent dna ligase from t.2 filiformis
|
| 82 | d2evra2 |
|
not modelled |
12.6 |
26 |
Fold:Cysteine proteinases
Superfamily:Cysteine proteinases
Family:NlpC/P60 |
| 83 | d2jfga2 |
|
not modelled |
12.5 |
27 |
Fold:MurD-like peptide ligases, peptide-binding domain
Superfamily:MurD-like peptide ligases, peptide-binding domain
Family:MurCDEF C-terminal domain |
| 84 | c3pjyB_ |
|
not modelled |
12.3 |
29 |
PDB header:transcription regulator
Chain: B: PDB Molecule:hypothetical signal peptide protein;
PDBTitle: crystal structure of a putative transcription regulator (r01717) from2 sinorhizobium meliloti 1021 at 1.55 a resolution
|
| 85 | d3d3ra1 |
|
not modelled |
12.2 |
20 |
Fold:OB-fold
Superfamily:HupF/HypC-like
Family:HupF/HypC-like |
| 86 | d1u0la1 |
|
not modelled |
12.1 |
28 |
Fold:OB-fold
Superfamily:Nucleic acid-binding proteins
Family:Cold shock DNA-binding domain-like |
| 87 | c2k1gA_ |
|
not modelled |
12.1 |
28 |
PDB header:lipoprotein
Chain: A: PDB Molecule:lipoprotein spr;
PDBTitle: solution nmr structure of lipoprotein spr from escherichia coli k12.2 northeast structural genomics target er541-37-162
|
| 88 | d1smxa_ |
|
not modelled |
12.0 |
29 |
Fold:OB-fold
Superfamily:Nucleic acid-binding proteins
Family:Cold shock DNA-binding domain-like |
| 89 | c2fqpD_ |
|
not modelled |
12.0 |
9 |
PDB header:metal binding protein
Chain: D: PDB Molecule:hypothetical protein bp2299;
PDBTitle: crystal structure of a cupin domain (bp2299) from bordetella pertussis2 tohama i at 1.80 a resolution
|
| 90 | d2gbsa1 |
|
not modelled |
11.8 |
20 |
Fold:PUA domain-like
Superfamily:PUA domain-like
Family:Atu2648/PH1033-like |
| 91 | c3anuA_ |
|
not modelled |
11.8 |
11 |
PDB header:lyase
Chain: A: PDB Molecule:d-serine dehydratase;
PDBTitle: crystal structure of d-serine dehydratase from chicken kidney
|
| 92 | d1uija1 |
|
not modelled |
11.6 |
18 |
Fold:Double-stranded beta-helix
Superfamily:RmlC-like cupins
Family:Germin/Seed storage 7S protein |
| 93 | c2e4fA_ |
|
not modelled |
11.2 |
26 |
PDB header:transport protein
Chain: A: PDB Molecule:g protein-activated inward rectifier potassium channel 2;
PDBTitle: crystal structure of the cytoplasmic domain of g-protein-gated inward2 rectifier potassium channel kir3.2
|
| 94 | d1xnea_ |
|
not modelled |
11.1 |
64 |
Fold:PUA domain-like
Superfamily:PUA domain-like
Family:ProFAR isomerase associated domain |
| 95 | c1qorA_ |
|
not modelled |
11.0 |
23 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:quinone oxidoreductase;
PDBTitle: crystal structure of escherichia coli quinone2 oxidoreductase complexed with nadph
|
| 96 | c3pbiA_ |
|
not modelled |
10.9 |
23 |
PDB header:hydrolase
Chain: A: PDB Molecule:invasion protein;
PDBTitle: structure of the peptidoglycan hydrolase ripb (rv1478) from2 mycobacterium tuberculosis at 1.6 resolution
|
| 97 | c3kgzA_ |
|
not modelled |
10.8 |
16 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:cupin 2 conserved barrel domain protein;
PDBTitle: crystal structure of a cupin 2 conserved barrel domain protein from2 rhodopseudomonas palustris
|
| 98 | d1rc6a_ |
|
not modelled |
10.4 |
17 |
Fold:Double-stranded beta-helix
Superfamily:RmlC-like cupins
Family:YlbA-like |
| 99 | d1e2wa2 |
|
not modelled |
10.4 |
39 |
Fold:Barrel-sandwich hybrid
Superfamily:Rudiment single hybrid motif
Family:Cytochrome f, small domain |