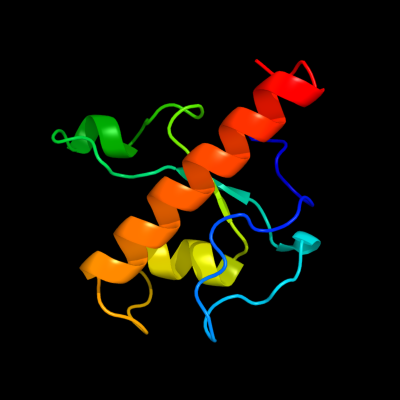
1 c3g27A_
100.0
99
PDB header: protein bindingChain: A: PDB Molecule: 82 prophage-derived uncharacterized protein ybco;PDBTitle: structure of a putative bacteriophage protein from escherichia coli2 str. k-12 substr. mg1655
2 d351ca_
40.5
23
Fold: Cytochrome cSuperfamily: Cytochrome cFamily: monodomain cytochrome c3 d1fi3a_
39.5
28
Fold: Cytochrome cSuperfamily: Cytochrome cFamily: monodomain cytochrome c4 c2d0sA_
34.6
26
PDB header: electron transportChain: A: PDB Molecule: cytochrome c;PDBTitle: crystal structure of the cytochrome c552 from moderate2 thermophilic bacterium, hydrogenophilus thermoluteolus
5 c2zxyA_
34.1
17
PDB header: oxygen binding, transport proteinChain: A: PDB Molecule: cytochrome c552;PDBTitle: crystal structure of cytochrome c555 from aquifex aeolicus
6 d1cora_
30.8
26
Fold: Cytochrome cSuperfamily: Cytochrome cFamily: monodomain cytochrome c7 d1dvva_
25.7
25
Fold: Cytochrome cSuperfamily: Cytochrome cFamily: monodomain cytochrome c8 d2izpa1
23.1
25
Fold: IpaD-likeSuperfamily: IpaD-likeFamily: IpaD-like9 d1ynra1
23.0
18
Fold: Cytochrome cSuperfamily: Cytochrome cFamily: monodomain cytochrome c10 c2izpB_
22.8
25
PDB header: toxinChain: B: PDB Molecule: putative membrane antigen;PDBTitle: bipd - an invasion prtein associated with the type-iii2 secretion system of burkholderia pseudomallei.
11 d1mz4a_
22.3
20
Fold: Cytochrome cSuperfamily: Cytochrome cFamily: monodomain cytochrome c12 d1f1ca_
17.2
27
Fold: Cytochrome cSuperfamily: Cytochrome cFamily: monodomain cytochrome c13 c3dr0B_
17.0
27
PDB header: electron transportChain: B: PDB Molecule: cytochrome c6;PDBTitle: structure of reduced cytochrome c6 from synechococcus sp.2 pcc 7002
14 d1a56a_
16.8
25
Fold: Cytochrome cSuperfamily: Cytochrome cFamily: monodomain cytochrome c15 d1qn2a_
16.2
27
Fold: Cytochrome cSuperfamily: Cytochrome cFamily: monodomain cytochrome c16 d2gc4d1
13.7
36
Fold: Cytochrome cSuperfamily: Cytochrome cFamily: monodomain cytochrome c17 d2i5nc1
12.8
31
Fold: Multiheme cytochromesSuperfamily: Multiheme cytochromesFamily: Photosynthetic reaction centre (cytochrome subunit)18 c2jblC_
12.8
31
PDB header: electron transportChain: C: PDB Molecule: photosynthetic reaction center cytochrome cPDBTitle: photosynthetic reaction center from blastochloris viridis
19 d1h1oa2
12.6
67
Fold: Cytochrome cSuperfamily: Cytochrome cFamily: Two-domain cytochrome c20 c2d0wA_
12.5
33
PDB header: electron transportChain: A: PDB Molecule: cytochrome cl;PDBTitle: crystal structure of cytochrome cl from hyphomicrobium2 denitrificans
21 d1fcdc1
not modelled
12.4
20
Fold: Cytochrome cSuperfamily: Cytochrome cFamily: Two-domain cytochrome c22 d1lmsa_
not modelled
12.3
27
Fold: Cytochrome cSuperfamily: Cytochrome cFamily: monodomain cytochrome c23 d1ql3a_
not modelled
12.1
42
Fold: Cytochrome cSuperfamily: Cytochrome cFamily: monodomain cytochrome c24 c1fcdD_
not modelled
11.9
20
PDB header: electron transport(flavocytochrome)Chain: D: PDB Molecule: flavocytochrome c sulfide dehydrogenasePDBTitle: the structure of flavocytochrome c sulfide dehydrogenase2 from a purple phototrophic bacterium chromatium vinosum at3 2.5 angstroms resolution
25 c1w5cT_
not modelled
11.7
21
PDB header: photosynthesisChain: T: PDB Molecule: cytochrome c-550;PDBTitle: photosystem ii from thermosynechococcus elongatus
26 d1gnta_
not modelled
11.3
35
Fold: Prismane protein-likeSuperfamily: Prismane protein-likeFamily: Hybrid cluster protein (prismane protein)27 d2ccya_
not modelled
11.2
67
Fold: Four-helical up-and-down bundleSuperfamily: CytochromesFamily: Cytochrome c'-like28 d1cpqa_
not modelled
11.1
83
Fold: Four-helical up-and-down bundleSuperfamily: CytochromesFamily: Cytochrome c'-like29 d1mqva_
not modelled
11.0
67
Fold: Four-helical up-and-down bundleSuperfamily: CytochromesFamily: Cytochrome c'-like30 d1iqca1
not modelled
11.0
57
Fold: Cytochrome cSuperfamily: Cytochrome cFamily: Di-heme cytochrome c peroxidase31 c3dmiA_
not modelled
10.9
29
PDB header: electron transportChain: A: PDB Molecule: cytochrome c6;PDBTitle: crystallization and structural analysis of cytochrome c62 from the diatom phaeodactylum tricornutum at 1.5 a3 resolution
32 d1gnla_
not modelled
10.9
35
Fold: Prismane protein-likeSuperfamily: Prismane protein-likeFamily: Hybrid cluster protein (prismane protein)33 d1f1fa_
not modelled
10.8
57
Fold: Cytochrome cSuperfamily: Cytochrome cFamily: monodomain cytochrome c34 c2zzsW_
not modelled
10.7
57
PDB header: electron transportChain: W: PDB Molecule: PDBTitle: crystal structure of cytochrome c554 from vibrio2 parahaemolyticus strain rimd2210633
35 d1gqaa_
not modelled
10.6
67
Fold: Four-helical up-and-down bundleSuperfamily: CytochromesFamily: Cytochrome c'-like36 d2j8wa1
not modelled
10.6
67
Fold: Four-helical up-and-down bundleSuperfamily: CytochromesFamily: Cytochrome c'-like37 c3oa8A_
not modelled
10.5
57
PDB header: heme-binding protein/heme-binding proteiChain: A: PDB Molecule: soxa;PDBTitle: diheme soxax
38 c1m70D_
not modelled
10.5
33
PDB header: electron transportChain: D: PDB Molecule: cytochrome c4;PDBTitle: crystal structure of oxidized recombinant cytochrome c4 from2 pseudomonas stutzeri
39 d1gksa_
not modelled
10.4
83
Fold: Cytochrome cSuperfamily: Cytochrome cFamily: monodomain cytochrome c40 d1e85a_
not modelled
10.2
83
Fold: Four-helical up-and-down bundleSuperfamily: CytochromesFamily: Cytochrome c'-like41 d1cyja_
not modelled
10.2
57
Fold: Cytochrome cSuperfamily: Cytochrome cFamily: monodomain cytochrome c42 d1e29a_
not modelled
10.2
20
Fold: Cytochrome cSuperfamily: Cytochrome cFamily: monodomain cytochrome c43 d1hzua1
not modelled
10.1
14
Fold: Cytochrome cSuperfamily: Cytochrome cFamily: N-terminal (heme c) domain of cytochrome cd1-nitrite reductase44 d2rdza1
not modelled
10.1
20
Fold: Multiheme cytochromesSuperfamily: Multiheme cytochromesFamily: Di-heme elbow motif45 d1cc5a_
not modelled
10.1
80
Fold: Cytochrome cSuperfamily: Cytochrome cFamily: monodomain cytochrome c46 d1ctja_
not modelled
10.0
57
Fold: Cytochrome cSuperfamily: Cytochrome cFamily: monodomain cytochrome c47 d1c6ra_
not modelled
9.9
57
Fold: Cytochrome cSuperfamily: Cytochrome cFamily: monodomain cytochrome c48 d1i8oa_
not modelled
9.8
27
Fold: Cytochrome cSuperfamily: Cytochrome cFamily: monodomain cytochrome c49 d1s05a_
not modelled
9.8
50
Fold: Four-helical up-and-down bundleSuperfamily: CytochromesFamily: Cytochrome c'-like50 c2hgoA_
not modelled
9.7
33
PDB header: toxinChain: A: PDB Molecule: cassiicolin;PDBTitle: nmr structure of cassiicolin
51 d1bbha_
not modelled
9.7
50
Fold: Four-helical up-and-down bundleSuperfamily: CytochromesFamily: Cytochrome c'-like52 d1c75a_
not modelled
9.7
43
Fold: Cytochrome cSuperfamily: Cytochrome cFamily: monodomain cytochrome c53 d1nmla1
not modelled
9.6
50
Fold: Cytochrome cSuperfamily: Cytochrome cFamily: Di-heme cytochrome c peroxidase54 d1h32a2
not modelled
9.4
23
Fold: Cytochrome cSuperfamily: Cytochrome cFamily: Di-heme cytochrome c SoxA55 d1dvha_
not modelled
9.3
60
Fold: Cytochrome cSuperfamily: Cytochrome cFamily: monodomain cytochrome c56 d1m70a2
not modelled
9.3
67
Fold: Cytochrome cSuperfamily: Cytochrome cFamily: Two-domain cytochrome c57 d1tbxa_
not modelled
9.1
38
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: F93-like58 d1gu2a_
not modelled
9.1
57
Fold: Cytochrome cSuperfamily: Cytochrome cFamily: monodomain cytochrome c59 d1kb0a1
not modelled
9.0
60
Fold: Cytochrome cSuperfamily: Cytochrome cFamily: Quinoprotein alcohol dehydrogenase, C-terminal domain60 d1c52a_
not modelled
9.0
60
Fold: Cytochrome cSuperfamily: Cytochrome cFamily: monodomain cytochrome c61 d1h1oa1
not modelled
8.9
60
Fold: Cytochrome cSuperfamily: Cytochrome cFamily: Two-domain cytochrome c62 c1h32A_
not modelled
8.8
43
PDB header: electron transferChain: A: PDB Molecule: diheme cytochrome c;PDBTitle: reduced soxax complex from rhodovulum sulfidophilum
63 d1nira1
not modelled
8.6
14
Fold: Cytochrome cSuperfamily: Cytochrome cFamily: N-terminal (heme c) domain of cytochrome cd1-nitrite reductase64 d1h9xa1
not modelled
8.6
60
Fold: Cytochrome cSuperfamily: Cytochrome cFamily: N-terminal (heme c) domain of cytochrome cd1-nitrite reductase65 d1gdva_
not modelled
8.5
57
Fold: Cytochrome cSuperfamily: Cytochrome cFamily: monodomain cytochrome c66 d1nmla2
not modelled
8.5
67
Fold: Cytochrome cSuperfamily: Cytochrome cFamily: Di-heme cytochrome c peroxidase67 c2zonG_
not modelled
8.5
50
PDB header: oxidoreductase/electron transportChain: G: PDB Molecule: cytochrome c551;PDBTitle: crystal structure of electron transfer complex of nitrite2 reductase with cytochrome c
68 c1w2lA_
not modelled
8.4
67
PDB header: oxidoreductaseChain: A: PDB Molecule: cytochrome oxidase subunit ii;PDBTitle: cytochrome c domain of caa3 oxygen oxidoreductase
69 d1hj3a1
not modelled
8.4
60
Fold: Cytochrome cSuperfamily: Cytochrome cFamily: N-terminal (heme c) domain of cytochrome cd1-nitrite reductase70 c1fs9A_
not modelled
8.3
16
PDB header: oxidoreductaseChain: A: PDB Molecule: cytochrome c nitrite reductase;PDBTitle: cytochrome c nitrite reductase from wolinella succinogenes-azide2 complex
71 c2c1dC_
not modelled
8.0
20
PDB header: oxidoreductaseChain: C: PDB Molecule: soxa;PDBTitle: crystal structure of soxxa from p. pantotrophus
72 d1wvec1
not modelled
8.0
60
Fold: Cytochrome cSuperfamily: Cytochrome cFamily: monodomain cytochrome c73 d1kv9a1
not modelled
8.0
80
Fold: Cytochrome cSuperfamily: Cytochrome cFamily: Quinoprotein alcohol dehydrogenase, C-terminal domain74 d1eb7a1
not modelled
7.9
50
Fold: Cytochrome cSuperfamily: Cytochrome cFamily: Di-heme cytochrome c peroxidase75 d1dw0a_
not modelled
7.8
43
Fold: Cytochrome cSuperfamily: Cytochrome cFamily: monodomain cytochrome c76 d1qksa1
not modelled
7.8
60
Fold: Cytochrome cSuperfamily: Cytochrome cFamily: N-terminal (heme c) domain of cytochrome cd1-nitrite reductase77 d1e2rb1
not modelled
7.8
60
Fold: Cytochrome cSuperfamily: Cytochrome cFamily: N-terminal (heme c) domain of cytochrome cd1-nitrite reductase78 d3cx5d1
not modelled
7.7
80
Fold: Cytochrome cSuperfamily: Cytochrome cFamily: Cytochrome bc1 domain79 d1kx7a_
not modelled
7.6
43
Fold: Cytochrome cSuperfamily: Cytochrome cFamily: monodomain cytochrome c80 d1wada_
not modelled
7.6
22
Fold: Multiheme cytochromesSuperfamily: Multiheme cytochromesFamily: Cytochrome c3-like81 c2fwtA_
not modelled
7.5
63
PDB header: electron transportChain: A: PDB Molecule: dhc, diheme cytochrome c;PDBTitle: crystal structure of dhc purified from rhodobacter2 sphaeroides
82 c3a1gD_
not modelled
7.5
29
PDB header: transferaseChain: D: PDB Molecule: polymerase basic protein 2;PDBTitle: high-resolution crystal structure of rna polymerase pb1-pb22 subunits from influenza a virus
83 c1pbyA_
not modelled
7.5
44
PDB header: oxidoreductaseChain: A: PDB Molecule: quinohemoprotein amine dehydrogenase 60 kdaPDBTitle: structure of the phenylhydrazine adduct of the2 quinohemoprotein amine dehydrogenase from paracoccus3 denitrificans at 1.7 a resolution
84 d1u5sb1
not modelled
7.5
45
Fold: Glucocorticoid receptor-like (DNA-binding domain)Superfamily: Glucocorticoid receptor-like (DNA-binding domain)Family: LIM domain85 d1dy7b1
not modelled
7.4
60
Fold: Cytochrome cSuperfamily: Cytochrome cFamily: N-terminal (heme c) domain of cytochrome cd1-nitrite reductase86 c1p84D_
not modelled
7.4
80
PDB header: oxidoreductaseChain: D: PDB Molecule: cytochrome c1, heme protein;PDBTitle: hdbt inhibited yeast cytochrome bc1 complex
87 c2yiuE_
not modelled
7.2
100
PDB header: oxidoreductaseChain: E: PDB Molecule: cytochrome c1, heme protein;PDBTitle: x-ray structure of the dimeric cytochrome bc1 complex from2 the soil bacterium paracoccus denitrificans at 2.73 angstrom resolution
88 d1fuia1
not modelled
7.2
28
Fold: Reductase/isomerase/elongation factor common domainSuperfamily: FucI/AraA C-terminal domain-likeFamily: L-fucose isomerase, C-terminal domain89 c3mk7B_
not modelled
7.2
50
PDB header: oxidoreductaseChain: B: PDB Molecule: cytochrome c oxidase, cbb3-type, subunit o;PDBTitle: the structure of cbb3 cytochrome oxidase
90 c1zzhA_
not modelled
7.2
43
PDB header: oxidoreductaseChain: A: PDB Molecule: cytochrome c peroxidase;PDBTitle: structure of the fully oxidized di-heme cytochrome c2 peroxidase from r. capsulatus
91 d1ppjd1
not modelled
7.0
80
Fold: Cytochrome cSuperfamily: Cytochrome cFamily: Cytochrome bc1 domain92 d1fs7a_
not modelled
7.0
16
Fold: Multiheme cytochromesSuperfamily: Multiheme cytochromesFamily: Di-heme elbow motif93 c3hq7A_
not modelled
6.9
57
PDB header: oxidoreductaseChain: A: PDB Molecule: cytochrome c551 peroxidase;PDBTitle: ccpa from g. sulfurreducens, g94k/k97q/r100i variant
94 d1c53a_
not modelled
6.9
50
Fold: Cytochrome cSuperfamily: Cytochrome cFamily: monodomain cytochrome c95 d1m70a1
not modelled
6.9
80
Fold: Cytochrome cSuperfamily: Cytochrome cFamily: Two-domain cytochrome c96 c2echA_
not modelled
6.9
30
PDB header: blood coagulation inhibitorChain: A: PDB Molecule: echistatin;PDBTitle: echistatin-the refined structure of a disintegrin in2 solution by 1h nmr
97 d1pbya1
not modelled
6.8
80
Fold: Cytochrome cSuperfamily: Cytochrome cFamily: Quinohemoprotein amine dehydrogenase A chain, domains 1 and 298 c2v07A_
not modelled
6.7
71
PDB header: photosynthesisChain: A: PDB Molecule: cytochrome c6;PDBTitle: structure of the arabidopsis thaliana cytochrome c6a v52q2 variant
99 c3cwbQ_
not modelled
6.6
100
PDB header: oxidoreductaseChain: Q: PDB Molecule: mitochondrial cytochrome c1, heme protein;PDBTitle: chicken cytochrome bc1 complex inhibited by an iodinated analogue of2 the polyketide crocacin-d