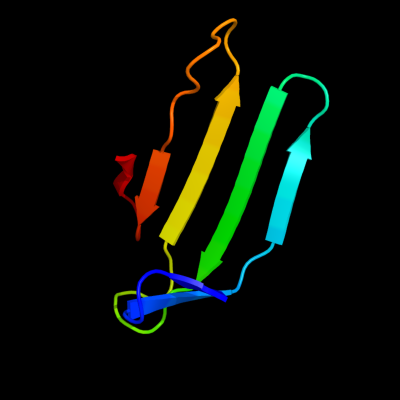
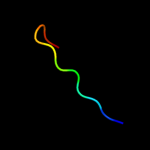
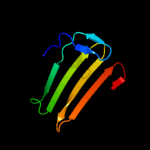
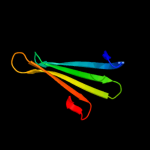
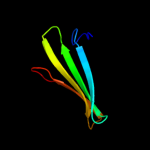
1 d1sa8a_
29.2
11
Fold: LipocalinsSuperfamily: LipocalinsFamily: Fatty acid binding protein-like2 d2cbra_
26.4
17
Fold: LipocalinsSuperfamily: LipocalinsFamily: Fatty acid binding protein-like3 d1hmsa_
26.3
19
Fold: LipocalinsSuperfamily: LipocalinsFamily: Fatty acid binding protein-like4 d2fs6a1
21.1
12
Fold: LipocalinsSuperfamily: LipocalinsFamily: Fatty acid binding protein-like5 d2r48a1
20.6
55
Fold: Phosphotyrosine protein phosphatases I-likeSuperfamily: PTS system IIB component-likeFamily: PTS system, Fructose specific IIB subunit-like6 c2gmhA_
19.3
17
PDB header: oxidoreductaseChain: A: PDB Molecule: electron transfer flavoprotein-ubiquinonePDBTitle: structure of porcine electron transfer flavoprotein-2 ubiquinone oxidoreductase in complexed with ubiquinone
7 c1u8cB_
19.1
15
PDB header: cell adhesionChain: B: PDB Molecule: integrin beta-3;PDBTitle: a novel adaptation of the integrin psi domain revealed from its2 crystal structure
8 c3k6sB_
18.3
15
PDB header: cell adhesionChain: B: PDB Molecule: integrin beta-2;PDBTitle: structure of integrin alphaxbeta2 ectodomain
9 d1hfel2
16.5
32
Fold: Ferredoxin-likeSuperfamily: 4Fe-4S ferredoxinsFamily: Ferredoxin domains from multidomain proteins10 c1nijA_
16.4
22
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: hypothetical protein yjia;PDBTitle: yjia protein
11 d1fcaa_
15.5
38
Fold: Ferredoxin-likeSuperfamily: 4Fe-4S ferredoxinsFamily: Short-chain ferredoxins12 d1blua_
15.2
44
Fold: Ferredoxin-likeSuperfamily: 4Fe-4S ferredoxinsFamily: Short-chain ferredoxins13 c2c3yA_
15.1
46
PDB header: oxidoreductaseChain: A: PDB Molecule: pyruvate-ferredoxin oxidoreductase;PDBTitle: crystal structure of the radical form of2 pyruvate:ferredoxin oxidoreductase from desulfovibrio3 africanus
14 c2kyrA_
15.1
45
PDB header: transferaseChain: A: PDB Molecule: fructose-like phosphotransferase enzyme iib component 1;PDBTitle: solution structure of enzyme iib subunit of pts system from2 escherichia coli k12. northeast structural genomics consortium target3 er315/ontario center for structural proteomics target ec0544
15 d1dura_
14.8
31
Fold: Ferredoxin-likeSuperfamily: 4Fe-4S ferredoxinsFamily: Short-chain ferredoxins16 d1lfoa_
14.6
25
Fold: LipocalinsSuperfamily: LipocalinsFamily: Fatty acid binding protein-like17 d1pmpa_
14.3
15
Fold: LipocalinsSuperfamily: LipocalinsFamily: Fatty acid binding protein-like18 d1yiva1
14.2
17
Fold: LipocalinsSuperfamily: LipocalinsFamily: Fatty acid binding protein-like19 d1bwya_
13.7
17
Fold: LipocalinsSuperfamily: LipocalinsFamily: Fatty acid binding protein-like20 d2hnxa1
13.6
8
Fold: LipocalinsSuperfamily: LipocalinsFamily: Fatty acid binding protein-like21 d1gtea5
not modelled
13.2
44
Fold: Ferredoxin-likeSuperfamily: 4Fe-4S ferredoxinsFamily: Ferredoxin domains from multidomain proteins22 c2jz2A_
not modelled
13.1
43
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: ssl0352 protein;PDBTitle: solution nmr structure of ssl0352 protein from synechocystis sp. pcc2 6803. northeast structural genomics consortium target sgr42
23 d1ifca_
not modelled
12.8
11
Fold: LipocalinsSuperfamily: LipocalinsFamily: Fatty acid binding protein-like24 c2lfeA_
not modelled
12.5
14
PDB header: ligaseChain: A: PDB Molecule: e3 ubiquitin-protein ligase hecw2;PDBTitle: solution nmr structure of n-terminal domain of human e3 ubiquitin-2 protein ligase hecw2, northeast structural genomics consortium (nesg)3 target ht6306a
25 d1kzwa_
not modelled
12.4
11
Fold: LipocalinsSuperfamily: LipocalinsFamily: Fatty acid binding protein-like26 d1kqwa_
not modelled
11.5
20
Fold: LipocalinsSuperfamily: LipocalinsFamily: Fatty acid binding protein-like27 c3fcuB_
not modelled
11.1
13
PDB header: cell adhesion/blood clottingChain: B: PDB Molecule: integrin beta-3;PDBTitle: structure of headpiece of integrin aiibb3 in open conformation
28 d2fdna_
not modelled
11.1
38
Fold: Ferredoxin-likeSuperfamily: 4Fe-4S ferredoxinsFamily: Short-chain ferredoxins29 d1fdqa_
not modelled
11.0
13
Fold: LipocalinsSuperfamily: LipocalinsFamily: Fatty acid binding protein-like30 d1jb0c_
not modelled
10.8
21
Fold: Ferredoxin-likeSuperfamily: 4Fe-4S ferredoxinsFamily: 7-Fe ferredoxin31 d2gpia1
not modelled
10.8
19
Fold: Shew3726-likeSuperfamily: Shew3726-likeFamily: Shew3726-like32 d1b56a_
not modelled
10.8
15
Fold: LipocalinsSuperfamily: LipocalinsFamily: Fatty acid binding protein-like33 d2r4qa1
not modelled
10.6
31
Fold: Phosphotyrosine protein phosphatases I-likeSuperfamily: PTS system IIB component-likeFamily: PTS system, Fructose specific IIB subunit-like34 d7fd1a_
not modelled
10.6
27
Fold: Ferredoxin-likeSuperfamily: 4Fe-4S ferredoxinsFamily: 7-Fe ferredoxin35 d1mdca_
not modelled
10.4
9
Fold: LipocalinsSuperfamily: LipocalinsFamily: Fatty acid binding protein-like36 c2fugG_
not modelled
10.1
18
PDB header: oxidoreductaseChain: G: PDB Molecule: nadh-quinone oxidoreductase chain 9;PDBTitle: crystal structure of the hydrophilic domain of respiratory complex i2 from thermus thermophilus
37 d1udxa3
not modelled
10.0
25
Fold: Obg GTP-binding protein C-terminal domainSuperfamily: Obg GTP-binding protein C-terminal domainFamily: Obg GTP-binding protein C-terminal domain38 d1gsma1
not modelled
10.0
26
Fold: Immunoglobulin-like beta-sandwichSuperfamily: ImmunoglobulinFamily: I set domains39 d2f1na1
not modelled
9.9
24
Fold: DNase I-likeSuperfamily: DNase I-likeFamily: DNase I-like40 c3dsoA_
not modelled
9.7
28
PDB header: metal binding proteinChain: A: PDB Molecule: putative uncharacterized protein copk;PDBTitle: crystal structure of cu(i) bound copper resistance protein copk
41 d2fug91
not modelled
9.6
18
Fold: Ferredoxin-likeSuperfamily: 4Fe-4S ferredoxinsFamily: Ferredoxin domains from multidomain proteins42 d1p6pa_
not modelled
9.6
18
Fold: LipocalinsSuperfamily: LipocalinsFamily: Fatty acid binding protein-like43 c2v4jE_
not modelled
9.6
16
PDB header: oxidoreductaseChain: E: PDB Molecule: sulfite reductase, dissimilatory-type subunitPDBTitle: the crystal structure of desulfovibrio vulgaris2 dissimilatory sulfite reductase bound to dsrc provides3 novel insights into the mechanism of sulfate respiration
44 c2q9sA_
not modelled
9.5
9
PDB header: lipid binding proteinChain: A: PDB Molecule: fatty acid-binding protein;PDBTitle: linoleic acid bound to fatty acid binding protein 4
45 c2lbaA_
not modelled
9.5
24
PDB header: lipid binding proteinChain: A: PDB Molecule: babp protein;PDBTitle: solution structure of chicken ileal babp in complex with2 glycochenodeoxycholic acid
46 d2f73a1
not modelled
9.1
26
Fold: LipocalinsSuperfamily: LipocalinsFamily: Fatty acid binding protein-like47 d1clfa_
not modelled
9.1
38
Fold: Ferredoxin-likeSuperfamily: 4Fe-4S ferredoxinsFamily: Short-chain ferredoxins48 c2zvsB_
not modelled
8.6
31
PDB header: electron transportChain: B: PDB Molecule: uncharacterized ferredoxin-like protein yfhl;PDBTitle: crystal structure of the 2[4fe-4s] ferredoxin from escherichia coli
49 d2c42a5
not modelled
8.5
25
Fold: Ferredoxin-likeSuperfamily: 4Fe-4S ferredoxinsFamily: Ferredoxin domains from multidomain proteins50 d2i9ua1
not modelled
8.5
21
Fold: Composite domain of metallo-dependent hydrolasesSuperfamily: Composite domain of metallo-dependent hydrolasesFamily: SAH/MTA deaminase-like51 c1gthD_
not modelled
8.5
44
PDB header: oxidoreductaseChain: D: PDB Molecule: dihydropyrimidine dehydrogenase;PDBTitle: dihydropyrimidine dehydrogenase (dpd) from pig, ternary2 complex with nadph and 5-iodouracil
52 d1vyfa_
not modelled
8.4
9
Fold: LipocalinsSuperfamily: LipocalinsFamily: Fatty acid binding protein-like53 c2vpyB_
not modelled
8.4
27
PDB header: oxidoreductaseChain: B: PDB Molecule: nrfc protein;PDBTitle: polysulfide reductase with bound quinone inhibitor,2 pentachlorophenol (pcp)
54 d1vlfn2
not modelled
8.1
38
Fold: Ferredoxin-likeSuperfamily: 4Fe-4S ferredoxinsFamily: Ferredoxin domains from multidomain proteins55 d1nekb1
not modelled
8.0
29
Fold: Globin-likeSuperfamily: alpha-helical ferredoxinFamily: Fumarate reductase/Succinate dehydogenase iron-sulfur protein, C-terminal domain56 c1gx7A_
not modelled
8.0
31
PDB header: oxidoreductaseChain: A: PDB Molecule: periplasmic [fe] hydrogenase large subunit;PDBTitle: best model of the electron transfer complex between2 cytochrome c3 and [fe]-hydrogenase
57 d1tw4a_
not modelled
7.9
20
Fold: LipocalinsSuperfamily: LipocalinsFamily: Fatty acid binding protein-like58 d1a57a_
not modelled
7.9
11
Fold: LipocalinsSuperfamily: LipocalinsFamily: Fatty acid binding protein-like59 d1g7na_
not modelled
7.7
13
Fold: LipocalinsSuperfamily: LipocalinsFamily: Fatty acid binding protein-like60 d3c7bb1
not modelled
7.7
25
Fold: Ferredoxin-likeSuperfamily: 4Fe-4S ferredoxinsFamily: Ferredoxin domains from multidomain proteins61 c2kxvA_
not modelled
7.7
19
PDB header: unknown functionChain: A: PDB Molecule: tellurite resistance protein;PDBTitle: nmr structure and calcium-binding properties of the tellurite2 resistance protein terd from klebsiella pneumoniae
62 c2vdcF_
not modelled
7.4
47
PDB header: oxidoreductaseChain: F: PDB Molecule: glutamate synthase [nadph] large chain;PDBTitle: the 9.5 a resolution structure of glutamate synthase from2 cryo-electron microscopy and its oligomerization behavior3 in solution: functional implications.
63 d1o1va_
not modelled
7.1
20
Fold: LipocalinsSuperfamily: LipocalinsFamily: Fatty acid binding protein-like64 d3c8ya3
not modelled
7.0
32
Fold: Ferredoxin-likeSuperfamily: 4Fe-4S ferredoxinsFamily: Ferredoxin domains from multidomain proteins65 d1h98a_
not modelled
6.9
31
Fold: Ferredoxin-likeSuperfamily: 4Fe-4S ferredoxinsFamily: 7-Fe ferredoxin66 c1lm1A_
not modelled
6.7
47
PDB header: oxidoreductaseChain: A: PDB Molecule: ferredoxin-dependent glutamate synthase;PDBTitle: structural studies on the synchronization of catalytic centers in2 glutamate synthase: native enzyme
67 c3pptA_
not modelled
6.7
17
PDB header: lipid binding proteinChain: A: PDB Molecule: sodium-calcium exchanger;PDBTitle: rep1-nxsq fatty acid transporter
68 c3v4pB_
not modelled
6.7
15
PDB header: cell adhesionChain: B: PDB Molecule: integrin beta-7;PDBTitle: crystal structure of a4b7 headpiece complexed with fab act-1
69 c3ef4A_
not modelled
6.6
14
PDB header: electron transportChain: A: PDB Molecule: blue copper protein;PDBTitle: crystal structure of native pseudoazurin from2 hyphomicrobium denitrificans
70 d1h0hb_
not modelled
6.5
13
Fold: Ferredoxin-likeSuperfamily: 4Fe-4S ferredoxinsFamily: Ferredoxin domains from multidomain proteins71 d1iqza_
not modelled
6.4
11
Fold: Ferredoxin-likeSuperfamily: 4Fe-4S ferredoxinsFamily: Single 4Fe-4S cluster ferredoxin72 c2bs2E_
not modelled
6.3
36
PDB header: oxidoreductaseChain: E: PDB Molecule: quinol-fumarate reductase iron-sulfur subunit b;PDBTitle: quinol:fumarate reductase from wolinella succinogenes
73 d1jnrb_
not modelled
6.1
20
Fold: Ferredoxin-likeSuperfamily: 4Fe-4S ferredoxinsFamily: Ferredoxin domains from multidomain proteins74 d2bs2b1
not modelled
6.0
36
Fold: Globin-likeSuperfamily: alpha-helical ferredoxinFamily: Fumarate reductase/Succinate dehydogenase iron-sulfur protein, C-terminal domain75 c3gyxJ_
not modelled
6.0
18
PDB header: oxidoreductaseChain: J: PDB Molecule: adenylylsulfate reductase;PDBTitle: crystal structure of adenylylsulfate reductase from2 desulfovibrio gigas
76 c3ibzA_
not modelled
5.9
22
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: putative tellurium resistant like protein terd;PDBTitle: crystal structure of putative tellurium resistant like protein (terd)2 from streptomyces coelicolor a3(2)
77 d2gmha3
not modelled
5.9
17
Fold: Ferredoxin-likeSuperfamily: 4Fe-4S ferredoxinsFamily: ETF-QO domain-like78 d1eala_
not modelled
5.8
22
Fold: LipocalinsSuperfamily: LipocalinsFamily: Fatty acid binding protein-like79 d1kf6b1
not modelled
5.7
29
Fold: Globin-likeSuperfamily: alpha-helical ferredoxinFamily: Fumarate reductase/Succinate dehydogenase iron-sulfur protein, C-terminal domain80 d1p9qc3
not modelled
5.7
29
Fold: Ferredoxin-likeSuperfamily: EF-G C-terminal domain-likeFamily: Hypothetical protein AF0491, C-terminal domain81 d1xera_
not modelled
5.6
31
Fold: Ferredoxin-likeSuperfamily: 4Fe-4S ferredoxinsFamily: Archaeal ferredoxins82 c1kqfB_
not modelled
5.6
40
PDB header: oxidoreductaseChain: B: PDB Molecule: formate dehydrogenase, nitrate-inducible, iron-sulfurPDBTitle: formate dehydrogenase n from e. coli
83 d1crba_
not modelled
5.5
19
Fold: LipocalinsSuperfamily: LipocalinsFamily: Fatty acid binding protein-like84 d2fug34
not modelled
5.4
29
Fold: Ferredoxin-likeSuperfamily: 4Fe-4S ferredoxinsFamily: Ferredoxin domains from multidomain proteins85 c2fgoA_
not modelled
5.3
44
PDB header: electron transportChain: A: PDB Molecule: ferredoxin;PDBTitle: structure of the 2[4fe-4s] ferredoxin from pseudomonas2 aeruginosa
86 d1rgva_
not modelled
5.3
31
Fold: Ferredoxin-likeSuperfamily: 4Fe-4S ferredoxinsFamily: Short-chain ferredoxins87 d1t95a3
not modelled
5.3
29
Fold: Ferredoxin-likeSuperfamily: EF-G C-terminal domain-likeFamily: Hypothetical protein AF0491, C-terminal domain88 c1ti2F_
not modelled
5.1
42
PDB header: oxidoreductaseChain: F: PDB Molecule: pyrogallol hydroxytransferase small subunit;PDBTitle: crystal structure of pyrogallol-phloroglucinol2 transhydroxylase from pelobacter acidigallici
89 c1dwlA_
not modelled
5.1
33
PDB header: electron transferChain: A: PDB Molecule: ferredoxin i;PDBTitle: the ferredoxin-cytochrome complex using heteronuclear nmr2 and docking simulation