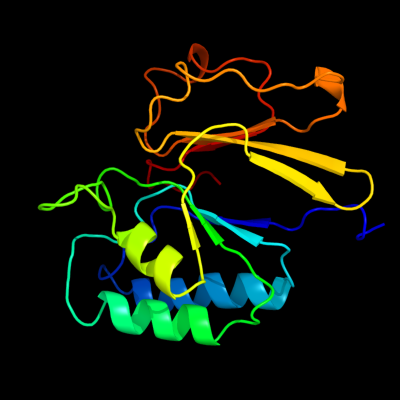
| 1 | c2xmoB_
|
|
 |
99.9 |
18 |
PDB header:hydrolase
Chain: B: PDB Molecule:lmo2642 protein;
PDBTitle: the crystal structure of lmo2642
|
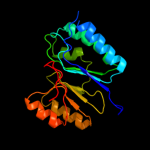
| 2 | c3av0A_
|
|
 |
99.9 |
15 |
PDB header:recombination
Chain: A: PDB Molecule:dna double-strand break repair protein mre11;
PDBTitle: crystal structure of mre11-rad50 bound to atp s
|
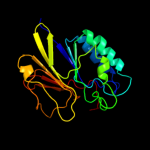
| 3 | c3ib7A_
|
|
 |
99.9 |
16 |
PDB header:hydrolase
Chain: A: PDB Molecule:icc protein;
PDBTitle: crystal structure of full length rv0805
|
| 4 | c3t1iC_
|
|
 |
99.9 |
25 |
PDB header:hydrolase
Chain: C: PDB Molecule:double-strand break repair protein mre11a;
PDBTitle: crystal structure of human mre11: understanding tumorigenic mutations
|
| 5 | d1xzwa2
|
|
 |
99.9 |
13 |
Fold:Metallo-dependent phosphatases
Superfamily:Metallo-dependent phosphatases
Family:Purple acid phosphatase-like |
| 6 | d3d03a1
|
|
 |
99.9 |
17 |
Fold:Metallo-dependent phosphatases
Superfamily:Metallo-dependent phosphatases
Family:GpdQ-like |
| 7 | d2yvta1
|
|
 |
99.9 |
20 |
Fold:Metallo-dependent phosphatases
Superfamily:Metallo-dependent phosphatases
Family:TT1561-like |
| 8 | c2hy1A_
|
|
 |
99.9 |
15 |
PDB header:hydrolase
Chain: A: PDB Molecule:rv0805;
PDBTitle: crystal structure of rv0805
|
| 9 | d2hy1a1
|
|
 |
99.9 |
15 |
Fold:Metallo-dependent phosphatases
Superfamily:Metallo-dependent phosphatases
Family:GpdQ-like |
| 10 | d2qfra2
|
|
 |
99.9 |
14 |
Fold:Metallo-dependent phosphatases
Superfamily:Metallo-dependent phosphatases
Family:Purple acid phosphatase-like |
| 11 | c3auzA_
|
|
 |
99.9 |
17 |
PDB header:recombination
Chain: A: PDB Molecule:dna double-strand break repair protein mre11;
PDBTitle: crystal structure of mre11 with manganese
|
| 12 | d1ii7a_
|
|
 |
99.8 |
21 |
Fold:Metallo-dependent phosphatases
Superfamily:Metallo-dependent phosphatases
Family:DNA double-strand break repair nuclease |
| 13 | c3qg5D_
|
|
 |
99.8 |
17 |
PDB header:hydrolase
Chain: D: PDB Molecule:mre11;
PDBTitle: the mre11:rad50 complex forms an atp dependent molecular clamp in dna2 double-strand break repair
|
| 14 | c1kbpB_
|
|
 |
99.8 |
14 |
PDB header:hydrolase (phosphoric monoester)
Chain: B: PDB Molecule:purple acid phosphatase;
PDBTitle: kidney bean purple acid phosphatase
|
| 15 | d1uf3a_
|
|
 |
99.8 |
18 |
Fold:Metallo-dependent phosphatases
Superfamily:Metallo-dependent phosphatases
Family:TT1561-like |
| 16 | c2q8uA_
|
|
 |
99.8 |
15 |
PDB header:hydrolase
Chain: A: PDB Molecule:exonuclease, putative;
PDBTitle: crystal structure of mre11 from thermotoga maritima msb8 (tm1635) at2 2.20 a resolution
|
| 17 | c1xzwB_
|
|
 |
99.8 |
14 |
PDB header:hydrolase
Chain: B: PDB Molecule:purple acid phosphatase;
PDBTitle: sweet potato purple acid phosphatase/phosphate complex
|
| 18 | c3rl4A_
|
|
 |
99.8 |
17 |
PDB header:hydrolase
Chain: A: PDB Molecule:metallophosphoesterase mpped2;
PDBTitle: rat metallophosphodiesterase mpped2 g252h mutant
|
| 19 | d2nxfa1
|
|
 |
99.7 |
21 |
Fold:Metallo-dependent phosphatases
Superfamily:Metallo-dependent phosphatases
Family:ADPRibase-Mn-like |
| 20 | d1s3la_
|
|
 |
99.7 |
23 |
Fold:Metallo-dependent phosphatases
Superfamily:Metallo-dependent phosphatases
Family:YfcE-like |
| 21 | c1s3mA_ |
|
not modelled |
99.7 |
23 |
PDB header:phosphodiesterase
Chain: A: PDB Molecule:hypothetical protein mj0936;
PDBTitle: structural and functional characterization of a novel2 archaeal phosphodiesterase
|
| 22 | d2z1aa2 |
|
not modelled |
99.7 |
16 |
Fold:Metallo-dependent phosphatases
Superfamily:Metallo-dependent phosphatases
Family:5'-nucleotidase (syn. UDP-sugar hydrolase), N-terminal domain |
| 23 | d1usha2 |
|
not modelled |
99.7 |
14 |
Fold:Metallo-dependent phosphatases
Superfamily:Metallo-dependent phosphatases
Family:5'-nucleotidase (syn. UDP-sugar hydrolase), N-terminal domain |
| 24 | d1utea_ |
|
not modelled |
99.7 |
20 |
Fold:Metallo-dependent phosphatases
Superfamily:Metallo-dependent phosphatases
Family:Purple acid phosphatase-like |
| 25 | c1qhwA_ |
|
not modelled |
99.7 |
17 |
PDB header:hydrolase
Chain: A: PDB Molecule:protein (purple acid phosphatase);
PDBTitle: purple acid phosphatase from rat bone
|
| 26 | d1qhwa_ |
|
not modelled |
99.7 |
17 |
Fold:Metallo-dependent phosphatases
Superfamily:Metallo-dependent phosphatases
Family:Purple acid phosphatase-like |
| 27 | c1oidA_ |
|
not modelled |
99.7 |
15 |
PDB header:hydrolase
Chain: A: PDB Molecule:protein usha;
PDBTitle: 5'-nucleotidase (e. coli) with an engineered disulfide2 bridge (s228c, p513c)
|
| 28 | c2z1aA_ |
|
not modelled |
99.7 |
19 |
PDB header:hydrolase
Chain: A: PDB Molecule:5'-nucleotidase;
PDBTitle: crystal structure of 5'-nucleotidase precursor from thermus2 thermophilus hb8
|
| 29 | c3qfnA_ |
|
not modelled |
99.6 |
20 |
PDB header:hydrolase
Chain: A: PDB Molecule:putative uncharacterized protein;
PDBTitle: crystal structure of streptococcal asymmetric ap4a hydrolase and2 phosphodiesterase spr1479/saph in complex with inorganic phosphate
|
| 30 | d2a22a1 |
|
not modelled |
99.6 |
20 |
Fold:Metallo-dependent phosphatases
Superfamily:Metallo-dependent phosphatases
Family:YfcE-like |
| 31 | d1z2wa1 |
|
not modelled |
99.6 |
23 |
Fold:Metallo-dependent phosphatases
Superfamily:Metallo-dependent phosphatases
Family:YfcE-like |
| 32 | c3rqzC_ |
|
not modelled |
99.5 |
15 |
PDB header:hydrolase
Chain: C: PDB Molecule:metallophosphoesterase;
PDBTitle: crystal structure of metallophosphoesterase from sphaerobacter2 thermophilus
|
| 33 | d1xm7a_ |
|
not modelled |
99.5 |
18 |
Fold:Metallo-dependent phosphatases
Superfamily:Metallo-dependent phosphatases
Family:Hypothetical protein aq 1666 |
| 34 | c3ivdA_ |
|
not modelled |
99.5 |
19 |
PDB header:hydrolase
Chain: A: PDB Molecule:nucleotidase;
PDBTitle: putative 5'-nucleotidase (c4898) from escherichia coli in2 complex with uridine
|
| 35 | c1su1A_ |
|
not modelled |
99.5 |
22 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:hypothetical protein yfce;
PDBTitle: structural and biochemical characterization of yfce, a2 phosphoesterase from e. coli
|
| 36 | d1su1a_ |
|
not modelled |
99.5 |
22 |
Fold:Metallo-dependent phosphatases
Superfamily:Metallo-dependent phosphatases
Family:YfcE-like |
| 37 | c3qfkA_ |
|
not modelled |
99.5 |
20 |
PDB header:hydrolase
Chain: A: PDB Molecule:uncharacterized protein;
PDBTitle: 2.05 angstrom crystal structure of putative 5'-nucleotidase from2 staphylococcus aureus in complex with alpha-ketoglutarate
|
| 38 | d3ck2a1 |
|
not modelled |
99.5 |
18 |
Fold:Metallo-dependent phosphatases
Superfamily:Metallo-dependent phosphatases
Family:YfcE-like |
| 39 | c3zu0A_ |
|
not modelled |
99.5 |
19 |
PDB header:hydrolase
Chain: A: PDB Molecule:nad nucleotidase;
PDBTitle: structure of haemophilus influenzae nad nucleotidase (nadn)
|
| 40 | c2wdfA_ |
|
not modelled |
99.4 |
21 |
PDB header:hydrolase
Chain: A: PDB Molecule:sulfur oxidation protein soxb;
PDBTitle: termus thermophilus sulfate thiohydrolase soxb
|
| 41 | d1nnwa_ |
|
not modelled |
99.4 |
13 |
Fold:Metallo-dependent phosphatases
Superfamily:Metallo-dependent phosphatases
Family:Phosphoesterase-related |
| 42 | c2kknA_ |
|
not modelled |
99.4 |
21 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:uncharacterized protein;
PDBTitle: solution nmr structure of themotoga maritima protein tm1076:2 northeast structural genomics consortium target vt57
|
| 43 | d3c9fa2 |
|
not modelled |
99.4 |
11 |
Fold:Metallo-dependent phosphatases
Superfamily:Metallo-dependent phosphatases
Family:5'-nucleotidase (syn. UDP-sugar hydrolase), N-terminal domain |
| 44 | c3gveB_ |
|
not modelled |
99.3 |
22 |
PDB header:structural genomics, unknown function
Chain: B: PDB Molecule:yfkn protein;
PDBTitle: crystal structure of calcineurin-like phosphoesterase yfkn from2 bacillus subtilis
|
| 45 | c3jyfB_ |
|
not modelled |
99.2 |
19 |
PDB header:hydrolase
Chain: B: PDB Molecule:2',3'-cyclic nucleotide 2'-phosphodiesterase/3'-
PDBTitle: the crystal structure of a 2,3-cyclic nucleotide 2-2 phosphodiesterase/3-nucleotidase bifunctional periplasmic precursor3 protein from klebsiella pneumoniae subsp. pneumoniae mgh 78578
|
| 46 | c3c9fB_ |
|
not modelled |
99.2 |
13 |
PDB header:hydrolase
Chain: B: PDB Molecule:5'-nucleotidase;
PDBTitle: crystal structure of 5'-nucleotidase from candida albicans sc5314
|
| 47 | d1t71a_ |
|
not modelled |
99.0 |
17 |
Fold:Metallo-dependent phosphatases
Superfamily:Metallo-dependent phosphatases
Family:DR1281-like |
| 48 | d2z06a1 |
|
not modelled |
98.8 |
16 |
Fold:Metallo-dependent phosphatases
Superfamily:Metallo-dependent phosphatases
Family:TTHA0625-like |
| 49 | d1t70a_ |
|
not modelled |
98.8 |
14 |
Fold:Metallo-dependent phosphatases
Superfamily:Metallo-dependent phosphatases
Family:DR1281-like |
| 50 | c2qjcA_ |
|
not modelled |
98.5 |
19 |
PDB header:hydrolase
Chain: A: PDB Molecule:diadenosine tetraphosphatase, putative;
PDBTitle: crystal structure of a putative diadenosine tetraphosphatase
|
| 51 | d1g5ba_ |
|
not modelled |
98.4 |
25 |
Fold:Metallo-dependent phosphatases
Superfamily:Metallo-dependent phosphatases
Family:Protein serine/threonine phosphatase |
| 52 | c2dfjA_ |
|
not modelled |
98.2 |
19 |
PDB header:hydrolase
Chain: A: PDB Molecule:diadenosinetetraphosphatase;
PDBTitle: crystal structure of the diadenosine tetraphosphate2 hydrolase from shigella flexneri 2a
|
| 53 | c2zbmA_ |
|
not modelled |
98.0 |
19 |
PDB header:hydrolase
Chain: A: PDB Molecule:protein-tyrosine-phosphatase;
PDBTitle: crystal structure of i115m mutant cold-active protein2 tyrosine phosphatase
|
| 54 | d1jk7a_ |
|
not modelled |
98.0 |
15 |
Fold:Metallo-dependent phosphatases
Superfamily:Metallo-dependent phosphatases
Family:Protein serine/threonine phosphatase |
| 55 | d3c5wc1 |
|
not modelled |
97.9 |
18 |
Fold:Metallo-dependent phosphatases
Superfamily:Metallo-dependent phosphatases
Family:Protein serine/threonine phosphatase |
| 56 | d1s70a_ |
|
not modelled |
97.8 |
15 |
Fold:Metallo-dependent phosphatases
Superfamily:Metallo-dependent phosphatases
Family:Protein serine/threonine phosphatase |
| 57 | c2jogA_ |
|
not modelled |
97.8 |
17 |
PDB header:hydrolase
Chain: A: PDB Molecule:calmodulin-dependent calcineurin a subunit alpha
PDBTitle: structure of the calcineurin-nfat complex
|
| 58 | c3icfB_ |
|
not modelled |
97.8 |
15 |
PDB header:hydrolase
Chain: B: PDB Molecule:serine/threonine-protein phosphatase t;
PDBTitle: structure of protein serine/threonine phosphatase from saccharomyces2 cerevisiae with similarity to human phosphatase pp5
|
| 59 | d1s95a_ |
|
not modelled |
97.8 |
16 |
Fold:Metallo-dependent phosphatases
Superfamily:Metallo-dependent phosphatases
Family:Protein serine/threonine phosphatase |
| 60 | c3e0jG_ |
|
not modelled |
97.8 |
14 |
PDB header:transferase
Chain: G: PDB Molecule:dna polymerase subunit delta-2;
PDBTitle: x-ray structure of the complex of regulatory subunits of2 human dna polymerase delta
|
| 61 | d1auia_ |
|
not modelled |
97.8 |
17 |
Fold:Metallo-dependent phosphatases
Superfamily:Metallo-dependent phosphatases
Family:Protein serine/threonine phosphatase |
| 62 | c1auiA_ |
|
not modelled |
97.8 |
17 |
PDB header:hydrolase
Chain: A: PDB Molecule:serine/threonine phosphatase 2b;
PDBTitle: human calcineurin heterodimer
|
| 63 | d2p6ba1 |
|
not modelled |
97.7 |
17 |
Fold:Metallo-dependent phosphatases
Superfamily:Metallo-dependent phosphatases
Family:Protein serine/threonine phosphatase |
| 64 | c2p6bC_ |
|
not modelled |
97.7 |
17 |
PDB header:hydrolase/hydrolase regulator
Chain: C: PDB Molecule:calmodulin-dependent calcineurin a subunit alpha
PDBTitle: crystal structure of human calcineurin in complex with2 pvivit peptide
|
| 65 | c1wao4_ |
|
not modelled |
97.5 |
16 |
PDB header:hydrolase
Chain: 4: PDB Molecule:serine/threonine protein phosphatase 5;
PDBTitle: pp5 structure
|
| 66 | c3floG_ |
|
not modelled |
97.1 |
18 |
PDB header:transferase
Chain: G: PDB Molecule:dna polymerase alpha subunit b;
PDBTitle: crystal structure of the carboxyl-terminal domain of yeast2 dna polymerase alpha in complex with its b subunit
|
| 67 | c2e76D_
|
|
 |
80.1 |
23 |
PDB header:photosynthesis
Chain: D: PDB Molecule:cytochrome b6-f complex iron-sulfur subunit;
PDBTitle: crystal structure of the cytochrome b6f complex with tridecyl-2 stigmatellin (tds) from m.laminosus
|
| 68 | c2fynO_ |
|
not modelled |
56.5 |
22 |
PDB header:oxidoreductase
Chain: O: PDB Molecule:ubiquinol-cytochrome c reductase iron-sulfur
PDBTitle: crystal structure analysis of the double mutant rhodobacter2 sphaeroides bc1 complex
|
| 69 | d3clsc1 |
|
not modelled |
56.4 |
11 |
Fold:Adenine nucleotide alpha hydrolase-like
Superfamily:Adenine nucleotide alpha hydrolases-like
Family:ETFP subunits |
| 70 | c1p84E_ |
|
not modelled |
54.9 |
14 |
PDB header:oxidoreductase
Chain: E: PDB Molecule:ubiquinol-cytochrome c reductase iron-sulfur
PDBTitle: hdbt inhibited yeast cytochrome bc1 complex
|
| 71 | c3ih5A_ |
|
not modelled |
50.5 |
21 |
PDB header:electron transport
Chain: A: PDB Molecule:electron transfer flavoprotein alpha-subunit;
PDBTitle: crystal structure of electron transfer flavoprotein alpha-2 subunit from bacteroides thetaiotaomicron
|
| 72 | d1efva1 |
|
not modelled |
48.5 |
21 |
Fold:Adenine nucleotide alpha hydrolase-like
Superfamily:Adenine nucleotide alpha hydrolases-like
Family:ETFP subunits |
| 73 | c1t9gR_ |
|
not modelled |
41.8 |
21 |
PDB header:oxidoreductase, electron transport
Chain: R: PDB Molecule:electron transfer flavoprotein alpha-subunit,
PDBTitle: structure of the human mcad:etf complex
|
| 74 | c2ebjB_ |
|
not modelled |
37.3 |
17 |
PDB header:hydrolase
Chain: B: PDB Molecule:pyrrolidone carboxyl peptidase;
PDBTitle: crystal structure of pyrrolidone carboxyl peptidase from thermus2 thermophilus
|
| 75 | c2fyuE_ |
|
not modelled |
35.0 |
15 |
PDB header:oxidoreductase
Chain: E: PDB Molecule:ubiquinol-cytochrome c reductase iron-sulfur subunit,
PDBTitle: crystal structure of bovine heart mitochondrial bc1 with jg1442 inhibitor
|
| 76 | d3bzka5 |
|
not modelled |
28.1 |
11 |
Fold:Ribonuclease H-like motif
Superfamily:Ribonuclease H-like
Family:Tex RuvX-like domain-like |
| 77 | d1kbia1 |
|
not modelled |
28.1 |
10 |
Fold:TIM beta/alpha-barrel
Superfamily:FMN-linked oxidoreductases
Family:FMN-linked oxidoreductases |
| 78 | d1o94c_ |
|
not modelled |
27.2 |
9 |
Fold:Adenine nucleotide alpha hydrolase-like
Superfamily:Adenine nucleotide alpha hydrolases-like
Family:ETFP subunits |
| 79 | c2ei9A_ |
|
not modelled |
27.0 |
13 |
PDB header:gene regulation
Chain: A: PDB Molecule:non-ltr retrotransposon r1bmks orf2 protein;
PDBTitle: crystal structure of r1bm endonuclease domain
|
| 80 | d1duvg2 |
|
not modelled |
26.2 |
14 |
Fold:ATC-like
Superfamily:Aspartate/ornithine carbamoyltransferase
Family:Aspartate/ornithine carbamoyltransferase |
| 81 | d1a2za_ |
|
not modelled |
21.9 |
17 |
Fold:Phosphorylase/hydrolase-like
Superfamily:Pyrrolidone carboxyl peptidase (pyroglutamate aminopeptidase)
Family:Pyrrolidone carboxyl peptidase (pyroglutamate aminopeptidase) |
| 82 | c3k2qA_ |
|
not modelled |
20.5 |
10 |
PDB header:transferase
Chain: A: PDB Molecule:pyrophosphate-dependent phosphofructokinase;
PDBTitle: crystal structure of pyrophosphate-dependent2 phosphofructokinase from marinobacter aquaeolei, northeast3 structural genomics consortium target mqr88
|
| 83 | d1uuya_ |
|
not modelled |
19.9 |
28 |
Fold:Molybdenum cofactor biosynthesis proteins
Superfamily:Molybdenum cofactor biosynthesis proteins
Family:MogA-like |
| 84 | c3rfqC_ |
|
not modelled |
19.6 |
20 |
PDB header:biosynthetic protein
Chain: C: PDB Molecule:pterin-4-alpha-carbinolamine dehydratase moab2;
PDBTitle: crystal structure of pterin-4-alpha-carbinolamine dehydratase moab22 from mycobacterium marinum
|
| 85 | d2f48a1 |
|
not modelled |
19.5 |
16 |
Fold:Phosphofructokinase
Superfamily:Phosphofructokinase
Family:Phosphofructokinase |
| 86 | d1auga_ |
|
not modelled |
18.8 |
19 |
Fold:Phosphorylase/hydrolase-like
Superfamily:Pyrrolidone carboxyl peptidase (pyroglutamate aminopeptidase)
Family:Pyrrolidone carboxyl peptidase (pyroglutamate aminopeptidase) |
| 87 | d1zl0a1 |
|
not modelled |
18.5 |
12 |
Fold:The "swivelling" beta/beta/alpha domain
Superfamily:LD-carboxypeptidase A C-terminal domain-like
Family:LD-carboxypeptidase A C-terminal domain-like |
| 88 | c3tebA_ |
|
not modelled |
18.3 |
19 |
PDB header:hydrolase
Chain: A: PDB Molecule:endonuclease/exonuclease/phosphatase;
PDBTitle: endonuclease/exonuclease/phosphatase family protein from leptotrichia2 buccalis c-1013-b
|
| 89 | d1dxha2 |
|
not modelled |
18.2 |
14 |
Fold:ATC-like
Superfamily:Aspartate/ornithine carbamoyltransferase
Family:Aspartate/ornithine carbamoyltransferase |
| 90 | c3qi7A_ |
|
not modelled |
17.9 |
17 |
PDB header:transcription
Chain: A: PDB Molecule:putative transcriptional regulator;
PDBTitle: crystal structure of a putative transcriptional regulator2 (yp_001089212.1) from clostridium difficile 630 at 1.86 a resolution
|
| 91 | c3n0vD_ |
|
not modelled |
17.3 |
16 |
PDB header:hydrolase
Chain: D: PDB Molecule:formyltetrahydrofolate deformylase;
PDBTitle: crystal structure of a formyltetrahydrofolate deformylase (pp_0327)2 from pseudomonas putida kt2440 at 2.25 a resolution
|
| 92 | c3obiC_ |
|
not modelled |
16.9 |
26 |
PDB header:hydrolase
Chain: C: PDB Molecule:formyltetrahydrofolate deformylase;
PDBTitle: crystal structure of a formyltetrahydrofolate deformylase (np_949368)2 from rhodopseudomonas palustris cga009 at 1.95 a resolution
|
| 93 | c3p9xB_ |
|
not modelled |
16.6 |
21 |
PDB header:transferase
Chain: B: PDB Molecule:phosphoribosylglycinamide formyltransferase;
PDBTitle: crystal structure of phosphoribosylglycinamide formyltransferase from2 bacillus halodurans
|
| 94 | c2higA_ |
|
not modelled |
16.4 |
11 |
PDB header:transferase
Chain: A: PDB Molecule:6-phospho-1-fructokinase;
PDBTitle: crystal structure of phosphofructokinase apoenzyme from trypanosoma2 brucei.
|
| 95 | c1cr6A_ |
|
not modelled |
16.3 |
13 |
PDB header:hydrolase
Chain: A: PDB Molecule:epoxide hydrolase;
PDBTitle: crystal structure of murine soluble epoxide hydrolase2 complexed with cpu inhibitor
|
| 96 | c2yxbA_ |
|
not modelled |
16.2 |
13 |
PDB header:isomerase
Chain: A: PDB Molecule:coenzyme b12-dependent mutase;
PDBTitle: crystal structure of the methylmalonyl-coa mutase alpha-subunit from2 aeropyrum pernix
|
| 97 | d1fh9a_ |
|
not modelled |
15.7 |
14 |
Fold:TIM beta/alpha-barrel
Superfamily:(Trans)glycosidases
Family:beta-glycanases |
| 98 | d1ua4a_ |
|
not modelled |
15.1 |
14 |
Fold:Ribokinase-like
Superfamily:Ribokinase-like
Family:ADP-specific Phosphofructokinase/Glucokinase |
| 99 | c3lacA_ |
|
not modelled |
14.9 |
26 |
PDB header:hydrolase
Chain: A: PDB Molecule:pyrrolidone-carboxylate peptidase;
PDBTitle: crystal structure of bacillus anthracis pyrrolidone-carboxylate2 peptidase, pcp
|