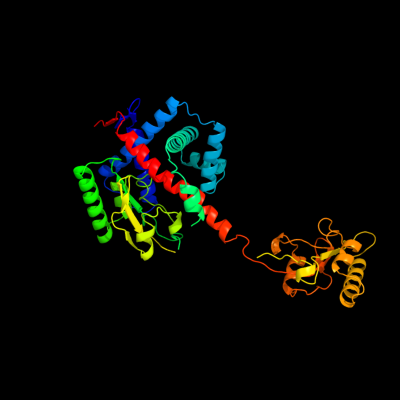
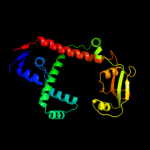
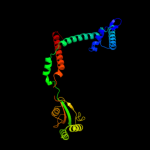
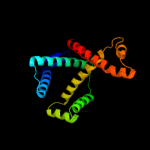
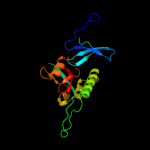
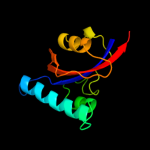
1 c1m5yB_
100.0
97
PDB header: isomerase, cell cycleChain: B: PDB Molecule: survival protein sura;PDBTitle: crystallographic structure of sura, a molecular chaperone2 that facilitates outer membrane porin folding
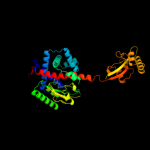
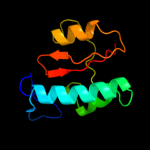
2 c2pv3B_
100.0
84
PDB header: isomeraseChain: B: PDB Molecule: chaperone sura;PDBTitle: crystallographic structure of sura fragment lacking the second2 peptidyl-prolyl isomerase domain complexed with peptide nftlkfwdifrk
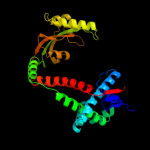
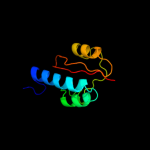
3 c3nrkA_
100.0
16
PDB header: unknown functionChain: A: PDB Molecule: lic12922;PDBTitle: the crystal structure of the leptospiral hypothetical protein lic12922
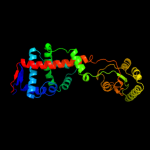
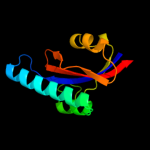
4 c3rgcB_
100.0
22
PDB header: chaperoneChain: B: PDB Molecule: possible periplasmic protein;PDBTitle: the virulence factor peb4 and the periplasmic protein cj1289 are two2 structurally related sura-like chaperones in the human pathogen3 campylobacter jejuni
5 c3rfwA_
100.0
25
PDB header: chaperoneChain: A: PDB Molecule: cell-binding factor 2;PDBTitle: the virulence factor peb4 and the periplasmic protein cj1289 are two2 structurally-related sura-like chaperones in the human pathogen3 campylobacter jejuni
6 d1m5ya1
99.9
86
Fold: Triger factor/SurA peptide-binding domain-likeSuperfamily: Triger factor/SurA peptide-binding domain-likeFamily: Porin chaperone SurA, peptide-binding domain7 c1f8aB_
99.8
23
PDB header: isomeraseChain: B: PDB Molecule: peptidyl-prolyl cis-trans isomerase nima-PDBTitle: structural basis for the phosphoserine-proline recognition2 by group iv ww domains
8 d1m5ya3
99.8
100
Fold: FKBP-likeSuperfamily: FKBP-likeFamily: FKBP immunophilin/proline isomerase9 c1yw5A_
99.8
21
PDB header: isomeraseChain: A: PDB Molecule: peptidyl prolyl cis/trans isomerase;PDBTitle: peptidyl-prolyl isomerase ess1 from candida albicans
10 c2jzvA_
99.7
33
PDB header: isomeraseChain: A: PDB Molecule: foldase protein prsa;PDBTitle: solution structure of s. aureus prsa-ppiase
11 d2pv2a1
99.7
100
Fold: FKBP-likeSuperfamily: FKBP-likeFamily: FKBP immunophilin/proline isomerase12 d1pina2
99.7
21
Fold: FKBP-likeSuperfamily: FKBP-likeFamily: FKBP immunophilin/proline isomerase13 d1j6ya_
99.6
26
Fold: FKBP-likeSuperfamily: FKBP-likeFamily: FKBP immunophilin/proline isomerase14 c3gpkA_
99.6
27
PDB header: isomeraseChain: A: PDB Molecule: ppic-type peptidyl-prolyl cis-trans isomerase;PDBTitle: crystal structure of ppic-type peptidyl-prolyl cis-trans isomerase2 domain at 1.55a resolution.
15 d1jnsa_
99.6
31
Fold: FKBP-likeSuperfamily: FKBP-likeFamily: FKBP immunophilin/proline isomerase16 c2rqsA_
99.6
37
PDB header: isomeraseChain: A: PDB Molecule: parvulin-like peptidyl-prolyl isomerase;PDBTitle: 3d structure of pin from the psychrophilic archeon cenarcheaum2 symbiosum (cspin)
17 d1eq3a_
99.5
35
Fold: FKBP-likeSuperfamily: FKBP-likeFamily: FKBP immunophilin/proline isomerase18 c2kgjA_
99.5
32
PDB header: isomeraseChain: A: PDB Molecule: peptidyl-prolyl cis-trans isomerase d;PDBTitle: solution structure of parvulin domain of ppid from e.coli
19 c1zk6A_
99.5
28
PDB header: isomeraseChain: A: PDB Molecule: foldase protein prsa;PDBTitle: nmr solution structure of b. subtilis prsa ppiase
20 c2nsaA_
97.3
15
PDB header: chaperoneChain: A: PDB Molecule: trigger factor;PDBTitle: structures of and interactions between domains of trigger factor from2 themotoga maritim
21 c3gtyX_
not modelled
97.0
13
PDB header: chaperone/ribosomal proteinChain: X: PDB Molecule: trigger factor;PDBTitle: promiscuous substrate recognition in folding and assembly activities2 of the trigger factor chaperone
22 d1w26a1
not modelled
96.8
15
Fold: Triger factor/SurA peptide-binding domain-likeSuperfamily: Triger factor/SurA peptide-binding domain-likeFamily: TF C-terminus23 c1w26B_
not modelled
96.2
12
PDB header: chaperoneChain: B: PDB Molecule: trigger factor;PDBTitle: trigger factor in complex with the ribosome forms a2 molecular cradle for nascent proteins
24 d1t11a1
not modelled
93.7
16
Fold: Triger factor/SurA peptide-binding domain-likeSuperfamily: Triger factor/SurA peptide-binding domain-likeFamily: TF C-terminus25 c1t11A_
not modelled
91.6
16
PDB header: chaperoneChain: A: PDB Molecule: trigger factor;PDBTitle: trigger factor
26 d1sknp_
not modelled
20.8
18
Fold: A DNA-binding domain in eukaryotic transcription factorsSuperfamily: A DNA-binding domain in eukaryotic transcription factorsFamily: A DNA-binding domain in eukaryotic transcription factors27 c2kz5A_
not modelled
17.8
16
PDB header: transcriptionChain: A: PDB Molecule: transcription factor nf-e2 45 kda subunit;PDBTitle: solution nmr structure of transcription factor nf-e2 subunit's dna2 binding domain from homo sapiens, northeast structural genomics3 consortium target hr4653b
28 d1rwsa_
not modelled
17.7
44
Fold: beta-Grasp (ubiquitin-like)Superfamily: MoaD/ThiSFamily: ThiS29 d1a75a_
not modelled
15.6
7
Fold: EF Hand-likeSuperfamily: EF-handFamily: Parvalbumin30 d1etob_
not modelled
11.1
19
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Homeodomain-likeFamily: FIS-like31 d1p5dx4
not modelled
10.7
14
Fold: TBP-likeSuperfamily: Phosphoglucomutase, C-terminal domainFamily: Phosphoglucomutase, C-terminal domain32 c2kycA_
not modelled
10.1
6
PDB header: calcium binding proteinChain: A: PDB Molecule: parvalbumin, thymic cpv3;PDBTitle: solution structure of ca-free chicken parvalbumin 3 (cpv3)
33 d1zata2
not modelled
8.8
15
Fold: L,D-transpeptidase pre-catalytic domain-likeSuperfamily: L,D-transpeptidase pre-catalytic domain-likeFamily: L,D-transpeptidase pre-catalytic domain-like34 c1q6uA_
not modelled
7.5
19
PDB header: isomeraseChain: A: PDB Molecule: fkbp-type peptidyl-prolyl cis-trans isomerase fkpa;PDBTitle: crystal structure of fkpa from escherichia coli
35 c3fs7D_
not modelled
7.3
6
PDB header: metal binding proteinChain: D: PDB Molecule: parvalbumin, thymic;PDBTitle: crystal structure of gallus gallus beta-parvalbumin (avian2 thymic hormone)
36 d1dq3a2
not modelled
7.0
9
Fold: dsRBD-likeSuperfamily: YcfA/nrd intein domainFamily: PI-Pfui intein middle domain37 d1stza1
not modelled
6.9
5
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: Heat-inducible transcription repressor HrcA, N-terminal domain38 d1bu3a_
not modelled
6.7
4
Fold: EF Hand-likeSuperfamily: EF-handFamily: Parvalbumin39 c3r9jD_
not modelled
6.6
17
PDB header: cell cycle,hydrolase/cell cycleChain: D: PDB Molecule: cell division topological specificity factor;PDBTitle: 4.3a resolution structure of a mind-mine(i24n) protein complex
40 c3c04A_
not modelled
6.6
10
PDB header: isomeraseChain: A: PDB Molecule: phosphomannomutase/phosphoglucomutase;PDBTitle: structure of the p368g mutant of pmm/pgm from p. aeruginosa
41 d1wdcb_
not modelled
6.5
9
Fold: EF Hand-likeSuperfamily: EF-handFamily: Calmodulin-like42 c3nr7A_
not modelled
6.3
23
PDB header: dna binding proteinChain: A: PDB Molecule: dna-binding protein h-ns;PDBTitle: crystal structure of s. typhimurium h-ns 1-83
43 d2jnaa1
not modelled
6.1
20
Fold: Dodecin subunit-likeSuperfamily: YdgH-likeFamily: YdgH-like44 d1u2ma_
not modelled
6.0
18
Fold: OmpH-likeSuperfamily: OmpH-likeFamily: OmpH-like45 d1rwya_
not modelled
5.9
9
Fold: EF Hand-likeSuperfamily: EF-handFamily: Parvalbumin46 d2pvba_
not modelled
5.8
7
Fold: EF Hand-likeSuperfamily: EF-handFamily: Parvalbumin47 d2jn6a1
not modelled
5.6
13
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Homeodomain-likeFamily: Cgl2762-like48 c3id5E_
not modelled
5.4
7
PDB header: transferase/ribosomal protein/rnaChain: E: PDB Molecule: pre mrna splicing protein;PDBTitle: crystal structure of sulfolobus solfataricus c/d rnp assembled with2 nop5, fibrillarin, l7ae and a split half c/d rna