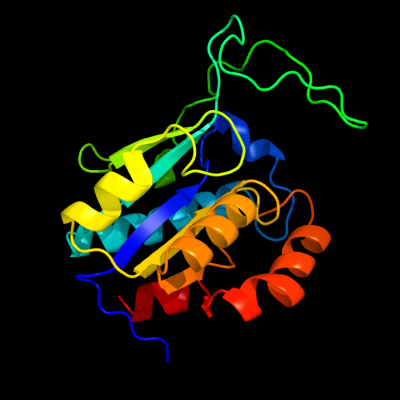
| 1 | d1j2ra_
|
|
 |
100.0 |
99 |
Fold:Isochorismatase-like hydrolases
Superfamily:Isochorismatase-like hydrolases
Family:Isochorismatase-like hydrolases |
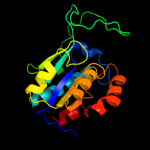
| 2 | d1nbaa_
|
|
 |
100.0 |
21 |
Fold:Isochorismatase-like hydrolases
Superfamily:Isochorismatase-like hydrolases
Family:Isochorismatase-like hydrolases |
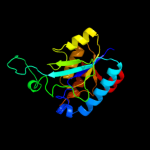
| 3 | c2fq1A_
|
|
 |
100.0 |
21 |
PDB header:hydrolase
Chain: A: PDB Molecule:isochorismatase;
PDBTitle: crystal structure of the two-domain non-ribosomal peptide synthetase2 entb containing isochorismate lyase and aryl-carrier protein domains
|
| 4 | c3irvA_
|
|
 |
100.0 |
24 |
PDB header:hydrolase
Chain: A: PDB Molecule:cysteine hydrolase;
PDBTitle: crystal structure of cysteine hydrolase pspph_2384 from pseudomonas2 syringae pv. phaseolicola 1448a
|
| 5 | c3kl2K_
|
|
 |
100.0 |
30 |
PDB header:structural genomics, unknown function
Chain: K: PDB Molecule:putative isochorismatase;
PDBTitle: crystal structure of a putative isochorismatase from streptomyces2 avermitilis
|
| 6 | d1nf9a_
|
|
 |
100.0 |
21 |
Fold:Isochorismatase-like hydrolases
Superfamily:Isochorismatase-like hydrolases
Family:Isochorismatase-like hydrolases |
| 7 | c3mcwA_
|
|
 |
100.0 |
30 |
PDB header:hydrolase
Chain: A: PDB Molecule:putative hydrolase;
PDBTitle: crystal structure of an a putative hydrolase of the isochorismatase2 family (cv_1320) from chromobacterium violaceum atcc 12472 at 1.06 a3 resolution
|
| 8 | c3hu5B_
|
|
 |
100.0 |
27 |
PDB header:hydrolase
Chain: B: PDB Molecule:isochorismatase family protein;
PDBTitle: crystal structure of isochorismatase family protein from desulfovibrio2 vulgaris subsp. vulgaris str. hildenborough
|
| 9 | c3ot4F_
|
|
 |
100.0 |
21 |
PDB header:hydrolase
Chain: F: PDB Molecule:putative isochorismatase;
PDBTitle: structure and catalytic mechanism of bordetella bronchiseptica nicf
|
| 10 | c3hb7G_
|
|
 |
100.0 |
21 |
PDB header:hydrolase
Chain: G: PDB Molecule:isochorismatase hydrolase;
PDBTitle: the crystal structure of an isochorismatase-like hydrolase from2 alkaliphilus metalliredigens to 2.3a
|
| 11 | c3oqpA_
|
|
 |
100.0 |
29 |
PDB header:hydrolase
Chain: A: PDB Molecule:putative isochorismatase;
PDBTitle: crystal structure of a putative isochorismatase (bxe_a0706) from2 burkholderia xenovorans lb400 at 1.22 a resolution
|
| 12 | c3oqpB_
|
|
 |
100.0 |
29 |
PDB header:hydrolase
Chain: B: PDB Molecule:putative isochorismatase;
PDBTitle: crystal structure of a putative isochorismatase (bxe_a0706) from2 burkholderia xenovorans lb400 at 1.22 a resolution
|
| 13 | c3eefA_
|
|
 |
100.0 |
27 |
PDB header:hydrolase
Chain: A: PDB Molecule:n-carbamoylsarcosine amidase related protein;
PDBTitle: crystal structure of n-carbamoylsarcosine amidase from thermoplasma2 acidophilum
|
| 14 | c3r2jC_
|
|
 |
100.0 |
16 |
PDB header:hydrolase
Chain: C: PDB Molecule:alpha/beta-hydrolase-like protein;
PDBTitle: crystal structure of pnc1 from l. infantum in complex with nicotinate
|
| 15 | c2wtaA_
|
|
 |
100.0 |
24 |
PDB header:hydrolase
Chain: A: PDB Molecule:nicotinamidase;
PDBTitle: acinetobacter baumanii nicotinamidase pyrazinamidease
|
| 16 | d1im5a_
|
|
 |
100.0 |
22 |
Fold:Isochorismatase-like hydrolases
Superfamily:Isochorismatase-like hydrolases
Family:Isochorismatase-like hydrolases |
| 17 | c3o93A_
|
|
 |
100.0 |
25 |
PDB header:hydrolase
Chain: A: PDB Molecule:nicotinamidase;
PDBTitle: high resolution crystal structures of streptococcus pneumoniae2 nicotinamidase with trapped intermediates provide insights into3 catalytic mechanism and inhibition by aldehydes
|
| 18 | c3lqyA_
|
|
 |
100.0 |
28 |
PDB header:hydrolase
Chain: A: PDB Molecule:putative isochorismatase hydrolase;
PDBTitle: crystal structure of putative isochorismatase hydrolase from2 oleispira antarctica
|
| 19 | d1yaca_
|
|
 |
100.0 |
18 |
Fold:Isochorismatase-like hydrolases
Superfamily:Isochorismatase-like hydrolases
Family:Isochorismatase-like hydrolases |
| 20 | c3gbcA_
|
|
 |
100.0 |
17 |
PDB header:hydrolase
Chain: A: PDB Molecule:pyrazinamidase/nicotinamidas pnca;
PDBTitle: determination of the crystal structure of the pyrazinamidase from2 m.tuberculosis : a structure-function analysis for prediction3 resistance to pyrazinamide
|
| 21 | c2a67C_ |
|
not modelled |
100.0 |
24 |
PDB header:structural genomics, unknown function
Chain: C: PDB Molecule:isochorismatase family protein;
PDBTitle: crystal structure of isochorismatase family protein
|
| 22 | c1yzvA_ |
|
not modelled |
100.0 |
19 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:hypothetical protein;
PDBTitle: hypothetical protein from trypanosoma cruzi
|
| 23 | c2b34C_ |
|
not modelled |
100.0 |
19 |
PDB header:hydrolase
Chain: C: PDB Molecule:mar1 ribonuclease;
PDBTitle: structure of mar1 ribonuclease from caenorhabditis elegans
|
| 24 | d1x9ga_ |
|
not modelled |
100.0 |
17 |
Fold:Isochorismatase-like hydrolases
Superfamily:Isochorismatase-like hydrolases
Family:Isochorismatase-like hydrolases |
| 25 | c2h0rD_ |
|
not modelled |
100.0 |
20 |
PDB header:hydrolase
Chain: D: PDB Molecule:nicotinamidase;
PDBTitle: structure of the yeast nicotinamidase pnc1p
|
| 26 | d1cmwa2 |
|
not modelled |
85.4 |
16 |
Fold:PIN domain-like
Superfamily:PIN domain-like
Family:5' to 3' exonuclease catalytic domain |
| 27 | c1x1qA_ |
|
not modelled |
80.8 |
17 |
PDB header:lyase
Chain: A: PDB Molecule:tryptophan synthase beta chain;
PDBTitle: crystal structure of tryptophan synthase beta chain from thermus2 thermophilus hb8
|
| 28 | d1tfra2 |
|
not modelled |
66.9 |
5 |
Fold:PIN domain-like
Superfamily:PIN domain-like
Family:5' to 3' exonuclease catalytic domain |
| 29 | c2ihnA_ |
|
not modelled |
66.7 |
7 |
PDB header:hydrolase/dna
Chain: A: PDB Molecule:ribonuclease h;
PDBTitle: co-crystal of bacteriophage t4 rnase h with a fork dna2 substrate
|
| 30 | c1ortD_ |
|
not modelled |
65.1 |
12 |
PDB header:transferase
Chain: D: PDB Molecule:ornithine transcarbamoylase;
PDBTitle: ornithine transcarbamoylase from pseudomonas aeruginosa
|
| 31 | d1xdpa3 |
|
not modelled |
65.0 |
9 |
Fold:Phospholipase D/nuclease
Superfamily:Phospholipase D/nuclease
Family:Polyphosphate kinase C-terminal domain |
| 32 | d1a9xa3 |
|
not modelled |
61.6 |
18 |
Fold:PreATP-grasp domain
Superfamily:PreATP-grasp domain
Family:BC N-terminal domain-like |
| 33 | c3bh0A_ |
|
not modelled |
60.7 |
15 |
PDB header:replication
Chain: A: PDB Molecule:dnab-like replicative helicase;
PDBTitle: atpase domain of g40p
|
| 34 | d1qopb_ |
|
not modelled |
57.8 |
18 |
Fold:Tryptophan synthase beta subunit-like PLP-dependent enzymes
Superfamily:Tryptophan synthase beta subunit-like PLP-dependent enzymes
Family:Tryptophan synthase beta subunit-like PLP-dependent enzymes |
| 35 | c1cmwA_ |
|
not modelled |
54.1 |
17 |
PDB header:transferase
Chain: A: PDB Molecule:protein (dna polymerase i);
PDBTitle: crystal structure of taq dna-polymerase shows a new2 orientation for the structure-specific nuclease domain
|
| 36 | d1vp8a_ |
|
not modelled |
48.2 |
19 |
Fold:Pyruvate kinase C-terminal domain-like
Superfamily:PK C-terminal domain-like
Family:MTH1675-like |
| 37 | c2w37A_ |
|
not modelled |
47.6 |
10 |
PDB header:transferase
Chain: A: PDB Molecule:ornithine carbamoyltransferase, catabolic;
PDBTitle: crystal structure of the hexameric catabolic ornithine2 transcarbamylase from lactobacillus hilgardii
|
| 38 | c1xdoB_ |
|
not modelled |
47.6 |
9 |
PDB header:transferase
Chain: B: PDB Molecule:polyphosphate kinase;
PDBTitle: crystal structure of escherichia coli polyphosphate kinase
|
| 39 | c1f8sA_ |
|
not modelled |
45.6 |
28 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:l-amino acid oxidase;
PDBTitle: crystal structure of l-amino acid oxidase from calloselasma2 rhodostoma, complexed with three molecules of o-aminobenzoate.
|
| 40 | c3bgwD_ |
|
not modelled |
43.4 |
15 |
PDB header:replication
Chain: D: PDB Molecule:dnab-like replicative helicase;
PDBTitle: the structure of a dnab-like replicative helicase and its interactions2 with primase
|
| 41 | c1vlvA_ |
|
not modelled |
43.0 |
12 |
PDB header:transferase
Chain: A: PDB Molecule:ornithine carbamoyltransferase;
PDBTitle: crystal structure of ornithine carbamoyltransferase (tm1097) from2 thermotoga maritima at 2.25 a resolution
|
| 42 | d1szpa2 |
|
not modelled |
42.1 |
11 |
Fold:P-loop containing nucleoside triphosphate hydrolases
Superfamily:P-loop containing nucleoside triphosphate hydrolases
Family:RecA protein-like (ATPase-domain) |
| 43 | c1xp8A_ |
|
not modelled |
40.0 |
17 |
PDB header:dna binding protein
Chain: A: PDB Molecule:reca protein;
PDBTitle: "deinococcus radiodurans reca in complex with atp-gamma-s"
|
| 44 | d2ae2a_ |
|
not modelled |
39.1 |
19 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Tyrosine-dependent oxidoreductases |
| 45 | d1pv8a_ |
|
not modelled |
36.0 |
18 |
Fold:TIM beta/alpha-barrel
Superfamily:Aldolase
Family:5-aminolaevulinate dehydratase, ALAD (porphobilinogen synthase) |
| 46 | c2rgoA_ |
|
not modelled |
35.3 |
21 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:alpha-glycerophosphate oxidase;
PDBTitle: structure of alpha-glycerophosphate oxidase from2 streptococcus sp.: a template for the mitochondrial alpha-3 glycerophosphate dehydrogenase
|
| 47 | c3cbwA_ |
|
not modelled |
34.4 |
8 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:ydht protein;
PDBTitle: crystal structure of the ydht protein from bacillus subtilis
|
| 48 | c3nugA_ |
|
not modelled |
32.4 |
21 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:3-oxoacyl-(acyl-carrier protein) reductase;
PDBTitle: crystal structure of wild type tetrameric pyridoxal 4-dehydrogenase2 from mesorhizobium loti
|
| 49 | c1ipaA_ |
|
not modelled |
31.8 |
18 |
PDB header:transferase
Chain: A: PDB Molecule:rna 2'-o-ribose methyltransferase;
PDBTitle: crystal structure of rna 2'-o ribose methyltransferase
|
| 50 | d1xp8a1 |
|
not modelled |
30.2 |
19 |
Fold:P-loop containing nucleoside triphosphate hydrolases
Superfamily:P-loop containing nucleoside triphosphate hydrolases
Family:RecA protein-like (ATPase-domain) |
| 51 | c2dflA_ |
|
not modelled |
29.9 |
10 |
PDB header:recombination
Chain: A: PDB Molecule:dna repair and recombination protein rada;
PDBTitle: crystal structure of left-handed rada filament
|
| 52 | d2iida1 |
|
not modelled |
29.0 |
24 |
Fold:FAD/NAD(P)-binding domain
Superfamily:FAD/NAD(P)-binding domain
Family:FAD-linked reductases, N-terminal domain |
| 53 | c1t4gA_ |
|
not modelled |
28.5 |
13 |
PDB header:recombination
Chain: A: PDB Molecule:dna repair and recombination protein rada;
PDBTitle: atpase in complex with amp-pnp
|
| 54 | d1duvg2 |
|
not modelled |
28.1 |
15 |
Fold:ATC-like
Superfamily:Aspartate/ornithine carbamoyltransferase
Family:Aspartate/ornithine carbamoyltransferase |
| 55 | d1ae1a_ |
|
not modelled |
28.0 |
11 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Tyrosine-dependent oxidoreductases |
| 56 | d1dxha2 |
|
not modelled |
27.2 |
17 |
Fold:ATC-like
Superfamily:Aspartate/ornithine carbamoyltransferase
Family:Aspartate/ornithine carbamoyltransferase |
| 57 | d1xsea_ |
|
not modelled |
26.9 |
23 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Tyrosine-dependent oxidoreductases |
| 58 | c1z2iA_ |
|
not modelled |
26.9 |
16 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:malate dehydrogenase;
PDBTitle: crystal structure of agrobacterium tumefaciens malate2 dehydrogenase, new york structural genomics consortium
|
| 59 | d1xu9a_ |
|
not modelled |
26.5 |
25 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Tyrosine-dependent oxidoreductases |
| 60 | c3o26A_ |
|
not modelled |
26.2 |
14 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:salutaridine reductase;
PDBTitle: the structure of salutaridine reductase from papaver somniferum.
|
| 61 | c2ha8A_ |
|
not modelled |
25.8 |
9 |
PDB header:rna binding protein
Chain: A: PDB Molecule:tar (hiv-1) rna loop binding protein;
PDBTitle: methyltransferase domain of human tar (hiv-1) rna binding2 protein 1
|
| 62 | c3nklA_ |
|
not modelled |
25.8 |
13 |
PDB header:oxidoreductase/lyase
Chain: A: PDB Molecule:udp-d-quinovosamine 4-dehydrogenase;
PDBTitle: crystal structure of udp-d-quinovosamine 4-dehydrogenase from vibrio2 fischeri
|
| 63 | c2vyeA_ |
|
not modelled |
25.4 |
22 |
PDB header:hydrolase
Chain: A: PDB Molecule:replicative dna helicase;
PDBTitle: crystal structure of the dnac-ssdna complex
|
| 64 | d1tyza_ |
|
not modelled |
25.4 |
22 |
Fold:Tryptophan synthase beta subunit-like PLP-dependent enzymes
Superfamily:Tryptophan synthase beta subunit-like PLP-dependent enzymes
Family:Tryptophan synthase beta subunit-like PLP-dependent enzymes |
| 65 | d1f2da_ |
|
not modelled |
25.3 |
18 |
Fold:Tryptophan synthase beta subunit-like PLP-dependent enzymes
Superfamily:Tryptophan synthase beta subunit-like PLP-dependent enzymes
Family:Tryptophan synthase beta subunit-like PLP-dependent enzymes |
| 66 | d1v8za1 |
|
not modelled |
25.0 |
16 |
Fold:Tryptophan synthase beta subunit-like PLP-dependent enzymes
Superfamily:Tryptophan synthase beta subunit-like PLP-dependent enzymes
Family:Tryptophan synthase beta subunit-like PLP-dependent enzymes |
| 67 | c2y8kA_ |
|
not modelled |
24.8 |
13 |
PDB header:hydrolase
Chain: A: PDB Molecule:carbohydrate binding family 6;
PDBTitle: structure of ctgh5-cbm6, an arabinoxylan-specific xylanase.
|
| 68 | c3g68A_ |
|
not modelled |
24.7 |
18 |
PDB header:isomerase
Chain: A: PDB Molecule:putative phosphosugar isomerase;
PDBTitle: crystal structure of a putative phosphosugar isomerase (cd3275) from2 clostridium difficile 630 at 1.80 a resolution
|
| 69 | c3ldaA_ |
|
not modelled |
24.7 |
12 |
PDB header:dna binding protein
Chain: A: PDB Molecule:dna repair protein rad51;
PDBTitle: yeast rad51 h352y filament interface mutant
|
| 70 | c3euaD_ |
|
not modelled |
24.5 |
9 |
PDB header:isomerase
Chain: D: PDB Molecule:putative fructose-aminoacid-6-phosphate deglycase;
PDBTitle: crystal structure of a putative phosphosugar isomerase (bsu32610) from2 bacillus subtilis at 1.90 a resolution
|
| 71 | c3fkjA_ |
|
not modelled |
24.4 |
7 |
PDB header:isomerase
Chain: A: PDB Molecule:putative phosphosugar isomerases;
PDBTitle: crystal structure of a putative phosphosugar isomerase (stm_0572) from2 salmonella typhimurium lt2 at 2.12 a resolution
|
| 72 | c2recB_ |
|
not modelled |
24.3 |
17 |
PDB header:helicase
PDB COMPND:
|
| 73 | d1ooea_ |
|
not modelled |
24.3 |
10 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Tyrosine-dependent oxidoreductases |
| 74 | c3ijrF_ |
|
not modelled |
24.2 |
20 |
PDB header:oxidoreductase
Chain: F: PDB Molecule:oxidoreductase, short chain dehydrogenase/reductase family;
PDBTitle: 2.05 angstrom resolution crystal structure of a short chain2 dehydrogenase from bacillus anthracis str. 'ames ancestor' in complex3 with nad+
|
| 75 | c2nm0B_ |
|
not modelled |
24.0 |
23 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:probable 3-oxacyl-(acyl-carrier-protein) reductase;
PDBTitle: crystal structure of sco1815: a beta-ketoacyl-acyl carrier protein2 reductase from streptomyces coelicolor a3(2)
|
| 76 | c3updA_ |
|
not modelled |
23.9 |
11 |
PDB header:transferase
Chain: A: PDB Molecule:ornithine carbamoyltransferase;
PDBTitle: 2.9 angstrom crystal structure of ornithine carbamoyltransferase2 (argf) from vibrio vulnificus
|
| 77 | c3uoeB_ |
|
not modelled |
23.8 |
10 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:dehydrogenase;
PDBTitle: the crystal structure of dehydrogenase from sinorhizobium meliloti
|
| 78 | d1qsga_ |
|
not modelled |
23.4 |
16 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Tyrosine-dependent oxidoreductases |
| 79 | c3toxG_ |
|
not modelled |
23.1 |
19 |
PDB header:oxidoreductase
Chain: G: PDB Molecule:short chain dehydrogenase;
PDBTitle: crystal structure of a short chain dehydrogenase in complex with2 nad(p) from sinorhizobium meliloti 1021
|
| 80 | c2b4qB_ |
|
not modelled |
22.8 |
25 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:rhamnolipids biosynthesis 3-oxoacyl-[acyl-
PDBTitle: pseudomonas aeruginosa rhlg/nadp active-site complex
|
| 81 | d1xrha_ |
|
not modelled |
22.6 |
8 |
Fold:L-sulfolactate dehydrogenase-like
Superfamily:L-sulfolactate dehydrogenase-like
Family:L-sulfolactate dehydrogenase-like |
| 82 | d1c0pa1 |
|
not modelled |
22.4 |
21 |
Fold:Nucleotide-binding domain
Superfamily:Nucleotide-binding domain
Family:D-aminoacid oxidase, N-terminal domain |
| 83 | d1onfa2 |
|
not modelled |
21.5 |
12 |
Fold:FAD/NAD(P)-binding domain
Superfamily:FAD/NAD(P)-binding domain
Family:FAD/NAD-linked reductases, N-terminal and central domains |
| 84 | c2rghA_ |
|
not modelled |
21.1 |
21 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:alpha-glycerophosphate oxidase;
PDBTitle: structure of alpha-glycerophosphate oxidase from2 streptococcus sp.: a template for the mitochondrial alpha-3 glycerophosphate dehydrogenase
|
| 85 | d2o8ra3 |
|
not modelled |
21.0 |
11 |
Fold:Phospholipase D/nuclease
Superfamily:Phospholipase D/nuclease
Family:Polyphosphate kinase C-terminal domain |
| 86 | d1reoa1 |
|
not modelled |
20.9 |
26 |
Fold:FAD/NAD(P)-binding domain
Superfamily:FAD/NAD(P)-binding domain
Family:FAD-linked reductases, N-terminal domain |
| 87 | c2e4gB_ |
|
not modelled |
20.4 |
15 |
PDB header:biosynthetic protein, flavoprotein
Chain: B: PDB Molecule:tryptophan halogenase;
PDBTitle: rebh with bound l-trp
|
| 88 | d1xg5a_ |
|
not modelled |
20.3 |
20 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Tyrosine-dependent oxidoreductases |
| 89 | d1xq1a_ |
|
not modelled |
20.0 |
15 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Tyrosine-dependent oxidoreductases |
| 90 | d2i1qa2 |
|
not modelled |
19.8 |
13 |
Fold:P-loop containing nucleoside triphosphate hydrolases
Superfamily:P-loop containing nucleoside triphosphate hydrolases
Family:RecA protein-like (ATPase-domain) |
| 91 | c2v1dA_ |
|
not modelled |
19.5 |
32 |
PDB header:oxidoreductase/repressor
Chain: A: PDB Molecule:lysine-specific histone demethylase 1;
PDBTitle: structural basis of lsd1-corest selectivity in histone h32 recognition
|
| 92 | c2xagA_ |
|
not modelled |
19.5 |
32 |
PDB header:transcription
Chain: A: PDB Molecule:lysine-specific histone demethylase 1;
PDBTitle: crystal structure of lsd1-corest in complex with para-bromo-2 (-)-trans-2-phenylcyclopropyl-1-amine
|
| 93 | d1nxua_ |
|
not modelled |
19.1 |
12 |
Fold:L-sulfolactate dehydrogenase-like
Superfamily:L-sulfolactate dehydrogenase-like
Family:L-sulfolactate dehydrogenase-like |
| 94 | d1a9xa4 |
|
not modelled |
19.1 |
18 |
Fold:PreATP-grasp domain
Superfamily:PreATP-grasp domain
Family:BC N-terminal domain-like |
| 95 | d1iz5a1 |
|
not modelled |
18.9 |
18 |
Fold:DNA clamp
Superfamily:DNA clamp
Family:DNA polymerase processivity factor |
| 96 | d1rfma_ |
|
not modelled |
18.5 |
20 |
Fold:L-sulfolactate dehydrogenase-like
Superfamily:L-sulfolactate dehydrogenase-like
Family:L-sulfolactate dehydrogenase-like |
| 97 | c2weuD_ |
|
not modelled |
18.3 |
14 |
PDB header:antifungal protein
Chain: D: PDB Molecule:tryptophan 5-halogenase;
PDBTitle: crystal structure of tryptophan 5-halogenase (pyrh) complex2 with substrate tryptophan
|
| 98 | d1plqa1 |
|
not modelled |
17.9 |
9 |
Fold:DNA clamp
Superfamily:DNA clamp
Family:DNA polymerase processivity factor |
| 99 | d2c07a1 |
|
not modelled |
17.5 |
9 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Tyrosine-dependent oxidoreductases |