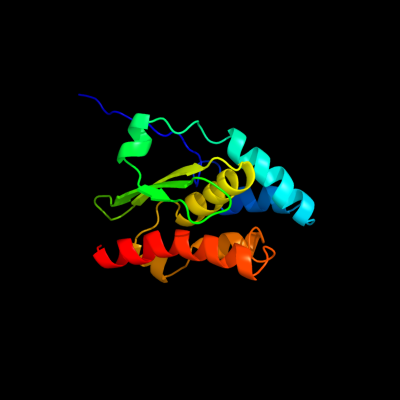
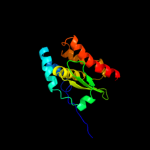
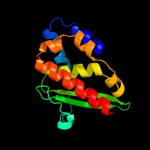
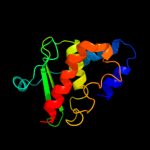
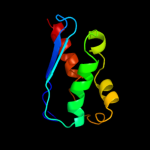
1 d1ni7a_
100.0
100
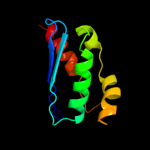
Fold: SufE/NifUSuperfamily: SufE/NifUFamily: SufE-like2 d1mzga_
100.0
33
Fold: SufE/NifUSuperfamily: SufE/NifUFamily: SufE-like3 c1wloA_
100.0
19
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: sufe protein;PDBTitle: solution structure of the hypothetical protein from thermus2 thermophilus hb8
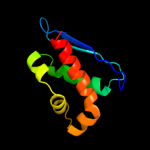
4 d1r9pa_
95.8
22
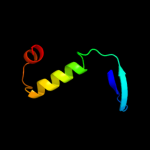
Fold: SufE/NifUSuperfamily: SufE/NifUFamily: NifU/IscU domain5 d1wfza_
91.7
19
Fold: SufE/NifUSuperfamily: SufE/NifUFamily: NifU/IscU domain6 c2z7eB_
83.7
19
PDB header: biosynthetic proteinChain: B: PDB Molecule: nifu-like protein;PDBTitle: crystal structure of aquifex aeolicus iscu with bound [2fe-2 2s] cluster
7 c2iy9A_
63.5
25
PDB header: toxinChain: A: PDB Molecule: suba;PDBTitle: crystal structure of the a-subunit of the ab5 toxin from e.2 coli
8 d1gcia_
62.8
15
Fold: Subtilisin-likeSuperfamily: Subtilisin-likeFamily: Subtilases9 c2b6nA_
62.0
18
PDB header: hydrolaseChain: A: PDB Molecule: proteinase k;PDBTitle: the 1.8 a crystal structure of a proteinase k like enzyme from a2 psychrotroph serratia species
10 d1r0re_
59.4
20
Fold: Subtilisin-likeSuperfamily: Subtilisin-likeFamily: Subtilases11 c1s2nB_
58.8
27
PDB header: hydrolaseChain: B: PDB Molecule: extracellular subtilisin-like serine proteinase;PDBTitle: crystal strucure of a cold adapted subtilisin-like serine proteinase
12 d2ixta1
55.3
20
Fold: Subtilisin-likeSuperfamily: Subtilisin-likeFamily: Subtilases13 c2w2qA_
54.4
35
PDB header: hydrolase/receptorChain: A: PDB Molecule: proprotein convertase subtilisin/kexin type 9;PDBTitle: pcsk9-deltac d374h mutant bound to wt egf-a of ldlr
14 c3f7oB_
53.4
20
PDB header: hydrolaseChain: B: PDB Molecule: serine protease;PDBTitle: crystal structure of cuticle-degrading protease from paecilomyces2 lilacinus (pl646)
15 c2oxaA_
52.0
19
PDB header: hydrolaseChain: A: PDB Molecule: extracellular serine protease;PDBTitle: crystal structure of serine protease of aeromonas sobria
16 d1gnsa_
51.3
18
Fold: Subtilisin-likeSuperfamily: Subtilisin-likeFamily: Subtilases17 d1p8ja2
50.2
23
Fold: Subtilisin-likeSuperfamily: Subtilisin-likeFamily: Subtilases18 d1to2e_
47.6
19
Fold: Subtilisin-likeSuperfamily: Subtilisin-likeFamily: Subtilases19 c2qq4A_
47.4
18
PDB header: metal binding proteinChain: A: PDB Molecule: iron-sulfur cluster biosynthesis protein iscu;PDBTitle: crystal structure of iron-sulfur cluster biosynthesis2 protein iscu (ttha1736) from thermus thermophilus hb8
20 d1v6ca_
45.9
15
Fold: Subtilisin-likeSuperfamily: Subtilisin-likeFamily: Subtilases21 d2id4a2
not modelled
45.4
23
Fold: Subtilisin-likeSuperfamily: Subtilisin-likeFamily: Subtilases22 d1pdaa2
not modelled
43.9
14
Fold: dsRBD-likeSuperfamily: Porphobilinogen deaminase (hydroxymethylbilane synthase), C-terminal domainFamily: Porphobilinogen deaminase (hydroxymethylbilane synthase), C-terminal domain23 c1r64A_
not modelled
43.1
23
PDB header: hydrolase/hydrolase inhibitorChain: A: PDB Molecule: kexin;PDBTitle: the 2.2 a crystal structure of kex2 protease in complex with ac-arg-2 glu-lys-boroarg peptidyl boronic acid inhibitor
24 c1p8jB_
not modelled
40.8
23
PDB header: hydrolase/hydrolase inhibitorChain: B: PDB Molecule: furin precursor;PDBTitle: crystal structure of the proprotein convertase furin
25 c3qfhE_
not modelled
40.4
18
PDB header: hydrolaseChain: E: PDB Molecule: epidermin leader peptide processing serine protease epip;PDBTitle: 2.05 angstrom resolution crystal structure of epidermin leader peptide2 processing serine protease (epip) from staphylococcus aureus.
26 d1gtka2
not modelled
39.1
14
Fold: dsRBD-likeSuperfamily: Porphobilinogen deaminase (hydroxymethylbilane synthase), C-terminal domainFamily: Porphobilinogen deaminase (hydroxymethylbilane synthase), C-terminal domain27 d1bh6a_
not modelled
38.7
19
Fold: Subtilisin-likeSuperfamily: Subtilisin-likeFamily: Subtilases28 c3lpcA_
not modelled
33.3
35
PDB header: hydrolaseChain: A: PDB Molecule: aprb2;PDBTitle: crystal structure of a subtilisin-like protease
29 c3bpsA_
not modelled
32.4
35
PDB header: hydrolase/lipid transportChain: A: PDB Molecule: proprotein convertase subtilisin/kexin type 9;PDBTitle: pcsk9:egf-a complex
30 d1nexa1
not modelled
31.1
26
Fold: Skp1 dimerisation domain-likeSuperfamily: Skp1 dimerisation domain-likeFamily: Skp1 dimerisation domain-like31 d1xjsa_
not modelled
27.0
18
Fold: SufE/NifUSuperfamily: SufE/NifUFamily: NifU/IscU domain32 d1fs1b1
not modelled
26.9
39
Fold: Skp1 dimerisation domain-likeSuperfamily: Skp1 dimerisation domain-likeFamily: Skp1 dimerisation domain-like33 c2yukA_
not modelled
25.6
14
PDB header: transferaseChain: A: PDB Molecule: myeloid/lymphoid or mixed-lineage leukemiaPDBTitle: solution structure of the hmg box of human myeloid/lymphoid2 or mixed-lineage leukemia protein 3 homolog
34 d1fs2b1
not modelled
25.5
39
Fold: Skp1 dimerisation domain-likeSuperfamily: Skp1 dimerisation domain-likeFamily: Skp1 dimerisation domain-like35 d1su0b_
not modelled
24.8
21
Fold: SufE/NifUSuperfamily: SufE/NifUFamily: NifU/IscU domain36 d2pwaa1
not modelled
23.5
23
Fold: Subtilisin-likeSuperfamily: Subtilisin-likeFamily: Subtilases37 d1r6va_
not modelled
22.2
23
Fold: Subtilisin-likeSuperfamily: Subtilisin-likeFamily: Subtilases38 d1meea_
not modelled
22.0
23
Fold: Subtilisin-likeSuperfamily: Subtilisin-likeFamily: Subtilases39 d2olra2
not modelled
18.8
8
Fold: PEP carboxykinase N-terminal domainSuperfamily: PEP carboxykinase N-terminal domainFamily: PEP carboxykinase N-terminal domain40 c2ypnA_
not modelled
18.1
14
PDB header: transferaseChain: A: PDB Molecule: protein (hydroxymethylbilane synthase);PDBTitle: hydroxymethylbilane synthase
41 d2ovra1
not modelled
17.8
39
Fold: Skp1 dimerisation domain-likeSuperfamily: Skp1 dimerisation domain-likeFamily: Skp1 dimerisation domain-like42 c3t41B_
not modelled
17.3
16
PDB header: hydrolaseChain: B: PDB Molecule: epidermin leader peptide processing serine protease epip;PDBTitle: 1.95 angstrom resolution crystal structure of epidermin leader peptide2 processing serine protease (epip) s393a mutant from staphylococcus3 aureus
43 d1go4a_
not modelled
17.3
12
Fold: The spindle assembly checkpoint protein mad2Superfamily: The spindle assembly checkpoint protein mad2Family: The spindle assembly checkpoint protein mad244 c3e21A_
not modelled
14.7
19
PDB header: apoptosisChain: A: PDB Molecule: fas-associated factor 1;PDBTitle: crystal structure of faf-1 uba domain
45 d1luca_
not modelled
13.9
14
Fold: TIM beta/alpha-barrelSuperfamily: Bacterial luciferase-likeFamily: Bacterial luciferase (alkanal monooxygenase)46 d2hh6a1
not modelled
13.8
33
Fold: Left-handed superhelixSuperfamily: BH3980-likeFamily: BH3980-like47 c3gq2B_
not modelled
13.7
20
PDB header: transport proteinChain: B: PDB Molecule: general vesicular transport factor p115;PDBTitle: crystal structure of the dimer of the p115 tether globular head domain
48 d1ii2a2
not modelled
12.4
20
Fold: PEP carboxykinase N-terminal domainSuperfamily: PEP carboxykinase N-terminal domainFamily: PEP carboxykinase N-terminal domain49 c3lysC_
not modelled
12.1
17
PDB header: recombinationChain: C: PDB Molecule: prophage pi2 protein 01, integrase;PDBTitle: crystal structure of the n-terminal domain of the prophage2 pi2 protein 01 (integrase) from lactococcus lactis,3 northeast structural genomics consortium target kr124f
50 d1dbia_
not modelled
11.9
25
Fold: Subtilisin-likeSuperfamily: Subtilisin-likeFamily: Subtilases51 c3i74B_
not modelled
10.7
15
PDB header: hydrolase/hydrolase inhibitorChain: B: PDB Molecule: subtilisin-like protease;PDBTitle: crystal structure of the plant subtilisin-like protease sbt3 in2 complex with a chloromethylketone inhibitor
52 c2w0gA_
not modelled
10.6
27
PDB header: chaperoneChain: A: PDB Molecule: hsp90 co-chaperone cdc37;PDBTitle: hsp90 co-chaperone cdc37
53 c2gjvF_
not modelled
10.3
19
PDB header: structural genomics, unknown functionChain: F: PDB Molecule: putative cytoplasmic protein;PDBTitle: crystal structure of a protein of unknown function from salmonella2 typhimurium
54 c2hl7A_
not modelled
10.1
23
PDB header: oxidoreductaseChain: A: PDB Molecule: cytochrome c-type biogenesis protein ccmh;PDBTitle: crystal structure of the periplasmic domain of ccmh from pseudomonas2 aeruginosa
55 d2gjva1
not modelled
10.1
19
Fold: Phage tail protein-likeSuperfamily: Phage tail protein-likeFamily: STM4215-like56 c2w3cA_
not modelled
10.1
20
PDB header: transport proteinChain: A: PDB Molecule: general vesicular transport factor p115;PDBTitle: globular head region of the human general vesicular2 transport factor p115
57 c2qk1A_
not modelled
10.0
7
PDB header: protein bindingChain: A: PDB Molecule: protein stu2;PDBTitle: structural basis of microtubule plus end tracking by2 xmap215, clip-170 and eb1
58 c2kkpA_
not modelled
9.8
15
PDB header: dna binding proteinChain: A: PDB Molecule: phage integrase;PDBTitle: solution nmr structure of the phage integrase sam-like2 domain from moth 1796 from moorella thermoacetica.3 northeast structural genomics consortium target mtr39k4 (residues 64-171).
59 c1bq0A_
not modelled
9.8
8
PDB header: chaperoneChain: A: PDB Molecule: dnaj;PDBTitle: j-domain (residues 1-77) of the escherichia coli n-terminal2 fragment (residues 1-104) of the molecular chaperone dnaj,3 nmr, 20 structures
60 d1w6ka2
not modelled
8.9
17
Fold: alpha/alpha toroidSuperfamily: Terpenoid cyclases/Protein prenyltransferasesFamily: Terpene synthases61 d1gm5a1
not modelled
8.9
22
Fold: Four-helical up-and-down bundleSuperfamily: RecG, N-terminal domainFamily: RecG, N-terminal domain62 d1n83a_
not modelled
8.6
6
Fold: Nuclear receptor ligand-binding domainSuperfamily: Nuclear receptor ligand-binding domainFamily: Nuclear receptor ligand-binding domain63 d1us7b_
not modelled
8.5
27
Fold: Hsp90 co-chaperone CDC37Superfamily: Hsp90 co-chaperone CDC37Family: Hsp90 co-chaperone CDC3764 c1us7B_
not modelled
8.5
27
PDB header: chaperoneChain: B: PDB Molecule: hsp90 co-chaperone cdc37;PDBTitle: complex of hsp90 and p50
65 c2kykA_
not modelled
8.5
29
PDB header: ligaseChain: A: PDB Molecule: e3 ubiquitin-protein ligase itchy homolog;PDBTitle: the sandwich region between two lmp2a py motif regulates the2 interaction between aip4ww2domain and py motif
66 c3lbdA_
not modelled
7.9
7
PDB header: nuclear receptorChain: A: PDB Molecule: retinoic acid receptor gamma;PDBTitle: ligand-binding domain of the human retinoic acid receptor2 gamma bound to 9-cis retinoic acid
67 c3bvoA_
not modelled
7.6
16
PDB header: chaperoneChain: A: PDB Molecule: co-chaperone protein hscb, mitochondrial precursor;PDBTitle: crystal structure of human co-chaperone protein hscb
68 c3dmbA_
not modelled
7.5
16
PDB header: oxidoreductaseChain: A: PDB Molecule: putative general stress protein 26 with a pnp-oxidase likePDBTitle: crystal structure of a putative general stress family protein2 (xcc2264) from xanthomonas campestris pv. campestris at 2.30 a3 resolution
69 c2oaxC_
not modelled
7.4
29
PDB header: transcriptionChain: C: PDB Molecule: mineralocorticoid receptor;PDBTitle: crystal structure of the s810l mutant mineralocorticoid2 receptor associated with sc9420
70 c2kvhA_
not modelled
7.2
24
PDB header: transcriptionChain: A: PDB Molecule: zinc finger and btb domain-containing protein 32;PDBTitle: structure of the three-cys2his2 domain of mouse testis zinc2 finger protein
71 c2dl0A_
not modelled
7.2
17
PDB header: signaling proteinChain: A: PDB Molecule: sam and sh3 domain-containing protein 1;PDBTitle: solution structure of the sam-domain of the sam and sh32 domain containing protein 1
72 d1fpoa1
not modelled
7.1
17
Fold: Long alpha-hairpinSuperfamily: Chaperone J-domainFamily: Chaperone J-domain73 d1x40a1
not modelled
7.1
13
Fold: SAM domain-likeSuperfamily: SAM/Pointed domainFamily: SAM (sterile alpha motif) domain74 d1xbla_
not modelled
6.9
8
Fold: Long alpha-hairpinSuperfamily: Chaperone J-domainFamily: Chaperone J-domain75 c3eq1A_
not modelled
6.7
22
PDB header: transferaseChain: A: PDB Molecule: porphobilinogen deaminase;PDBTitle: the crystal structure of human porphobilinogen deaminase at2 2.8a resolution
76 d1coka_
not modelled
6.7
27
Fold: SAM domain-likeSuperfamily: SAM/Pointed domainFamily: SAM (sterile alpha motif) domain77 c2kg5A_
not modelled
6.7
16
PDB header: signaling proteinChain: A: PDB Molecule: arf-gap, rho-gap domain, ank repeat and phPDBTitle: nmr solution structure of arap3-sam
78 d1dd4c_
not modelled
6.6
21
Fold: Ribosomal protein L7/12, oligomerisation (N-terminal) domainSuperfamily: Ribosomal protein L7/12, oligomerisation (N-terminal) domainFamily: Ribosomal protein L7/12, oligomerisation (N-terminal) domain79 d2fiqa1
not modelled
6.5
22
Fold: TIM beta/alpha-barrelSuperfamily: AldolaseFamily: GatZ-like80 d1dd3a1
not modelled
6.5
21
Fold: Ribosomal protein L7/12, oligomerisation (N-terminal) domainSuperfamily: Ribosomal protein L7/12, oligomerisation (N-terminal) domainFamily: Ribosomal protein L7/12, oligomerisation (N-terminal) domain81 d1pq9a_
not modelled
6.4
12
Fold: Nuclear receptor ligand-binding domainSuperfamily: Nuclear receptor ligand-binding domainFamily: Nuclear receptor ligand-binding domain82 d1dd4d_
not modelled
6.2
21
Fold: Ribosomal protein L7/12, oligomerisation (N-terminal) domainSuperfamily: Ribosomal protein L7/12, oligomerisation (N-terminal) domainFamily: Ribosomal protein L7/12, oligomerisation (N-terminal) domain83 d1i36a1
not modelled
6.1
20
Fold: 6-phosphogluconate dehydrogenase C-terminal domain-likeSuperfamily: 6-phosphogluconate dehydrogenase C-terminal domain-likeFamily: Conserved hypothetical protein MTH174784 c2kd1A_
not modelled
6.1
8
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: dna integration/recombination/invertion protein;PDBTitle: solution nmr structure of the integrase-like domain from2 bacillus cereus ordered locus bc_1272. northeast3 structural genomics consortium target bcr268f
85 c2kw0A_
not modelled
6.0
27
PDB header: oxidoreductaseChain: A: PDB Molecule: ccmh protein;PDBTitle: solution structure of n-terminal domain of ccmh from escherichia.coli
86 d1hdja_
not modelled
5.9
9
Fold: Long alpha-hairpinSuperfamily: Chaperone J-domainFamily: Chaperone J-domain87 d1sgga_
not modelled
5.8
13
Fold: SAM domain-likeSuperfamily: SAM/Pointed domainFamily: SAM (sterile alpha motif) domain88 d1oxja1
not modelled
5.7
7
Fold: SAM domain-likeSuperfamily: SAM/Pointed domainFamily: SAM (sterile alpha motif) domain89 c2wwaJ_
not modelled
5.7
33
PDB header: ribosomeChain: J: PDB Molecule: 60s ribosomal protein l19;PDBTitle: cryo-em structure of idle yeast ssh1 complex bound to the2 yeast 80s ribosome
90 c3bgdB_
not modelled
5.6
19
PDB header: transferaseChain: B: PDB Molecule: thiopurine s-methyltransferase;PDBTitle: thiopurine s-methyltransferase
91 c3zqpB_
not modelled
5.5
23
PDB header: dna-binding proteinChain: B: PDB Molecule: terminase small subunit;PDBTitle: crystal structure of the small terminase oligomerization2 domain from a spp1-like bacteriophage
92 d1eg3a2
not modelled
5.4
60
Fold: EF Hand-likeSuperfamily: EF-handFamily: EF-hand modules in multidomain proteins93 c1z5xU_
not modelled
5.4
20
PDB header: hormone/growth factor receptorChain: U: PDB Molecule: ultraspiracle protein (usp) a homologue of rxr;PDBTitle: hemipteran ecdysone receptor ligand-binding domain2 complexed with ponasterone a
94 d1thma_
not modelled
5.4
25
Fold: Subtilisin-likeSuperfamily: Subtilisin-likeFamily: Subtilases95 c3c18B_
not modelled
5.3
14
PDB header: transferaseChain: B: PDB Molecule: nucleotidyltransferase-like protein;PDBTitle: crystal structure of nucleotidyltransferase-like protein2 (zp_00538802.1) from exiguobacterium sibiricum 255-15 at 1.90 a3 resolution
96 c2eamA_
not modelled
5.3
13
PDB header: signaling proteinChain: A: PDB Molecule: putative 47 kda protein;PDBTitle: solution structure of the n-terminal sam-domain of a human2 putative 47 kda protein
97 c2khqA_
not modelled
5.3
10
PDB header: dna binding proteinChain: A: PDB Molecule: integrase;PDBTitle: solution nmr structure of a phage integrase ssp19472 fragment 59-159 from staphylococcus saprophyticus,3 northeast structural genomics consortium target syr103b
98 c3lccA_
not modelled
5.2
21
PDB header: transferaseChain: A: PDB Molecule: putative methyl chloride transferase;PDBTitle: structure of a sam-dependent halide methyltransferase from arabidopsis2 thaliana
99 d1r4va_
not modelled
5.1
21
Fold: Histone-foldSuperfamily: Histone-foldFamily: Bacterial histone-fold protein