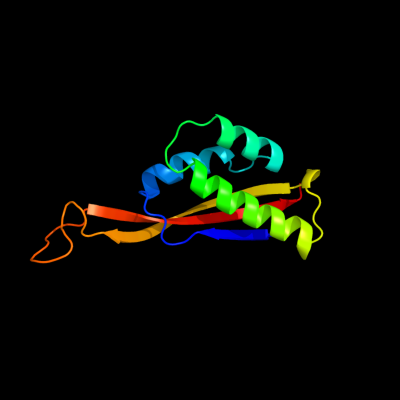
1 d2zjrp1
100.0
49
Fold: Ribosomal protein L22Superfamily: Ribosomal protein L22Family: Ribosomal protein L222 d1i4ja_
100.0
57
Fold: Ribosomal protein L22Superfamily: Ribosomal protein L22Family: Ribosomal protein L223 c3bboU_
100.0
36
PDB header: ribosomeChain: U: PDB Molecule: ribosomal protein l22;PDBTitle: homology model for the spinach chloroplast 50s subunit2 fitted to 9.4a cryo-em map of the 70s chlororibosome
4 d1vqor1
100.0
24
Fold: Ribosomal protein L22Superfamily: Ribosomal protein L22Family: Ribosomal protein L225 c4a17Q_
100.0
22
PDB header: ribosomeChain: Q: PDB Molecule: rpl17;PDBTitle: t.thermophila 60s ribosomal subunit in complex with2 initiation factor 6. this file contains 5s rrna,3 5.8s rrna and proteins of molecule 2.
6 c3iz5V_
100.0
30
PDB header: ribosomeChain: V: PDB Molecule: 60s ribosomal protein l17 (l22p);PDBTitle: localization of the large subunit ribosomal proteins into a 5.5 a2 cryo-em map of triticum aestivum translating 80s ribosome
7 c3jywN_
100.0
34
PDB header: ribosomeChain: N: PDB Molecule: 60s ribosomal protein l17(a);PDBTitle: structure of the 60s proteins for eukaryotic ribosome based on cryo-em2 map of thermomyces lanuginosus ribosome at 8.9a resolution
8 d2gycq1
100.0
100
Fold: Ribosomal protein L22Superfamily: Ribosomal protein L22Family: Ribosomal protein L229 c2ftcM_
100.0
29
PDB header: ribosomeChain: M: PDB Molecule: mitochondrial ribosomal protein l22 isoform a;PDBTitle: structural model for the large subunit of the mammalian mitochondrial2 ribosome
10 c2zkrr_
100.0
30
PDB header: ribosomal protein/rnaChain: R: PDB Molecule: rna expansion segment es39 part i;PDBTitle: structure of a mammalian ribosomal 60s subunit within an2 80s complex obtained by docking homology models of the rna3 and proteins into an 8.7 a cryo-em map
11 c1s1iN_
100.0
34
PDB header: ribosomeChain: N: PDB Molecule: 60s ribosomal protein l17-a;PDBTitle: structure of the ribosomal 80s-eef2-sordarin complex from2 yeast obtained by docking atomic models for rna and protein3 components into a 11.7 a cryo-em map. this file, 1s1i,4 contains 60s subunit. the 40s ribosomal subunit is in file5 1s1h.
12 d1r9pa_
85.9
20
Fold: SufE/NifUSuperfamily: SufE/NifUFamily: NifU/IscU domain13 c2z7eB_
74.8
19
PDB header: biosynthetic proteinChain: B: PDB Molecule: nifu-like protein;PDBTitle: crystal structure of aquifex aeolicus iscu with bound [2fe-2 2s] cluster
14 d1wfza_
67.9
21
Fold: SufE/NifUSuperfamily: SufE/NifUFamily: NifU/IscU domain15 c3ka5A_
32.6
13
PDB header: chaperoneChain: A: PDB Molecule: ribosome-associated protein y (psrp-1);PDBTitle: crystal structure of ribosome-associated protein y (psrp-1)2 from clostridium acetobutylicum. northeast structural3 genomics consortium target id car123a
16 d1fs1b1
29.6
33
Fold: Skp1 dimerisation domain-likeSuperfamily: Skp1 dimerisation domain-likeFamily: Skp1 dimerisation domain-like17 c3lyvF_
28.9
13
PDB header: chaperoneChain: F: PDB Molecule: ribosome-associated factor y;PDBTitle: crystal structure of a domain of ribosome-associated factor y from2 streptococcus pyogenes serotype m6. northeast structural genomics3 consortium target id dr64a
18 d2ouxa1
27.4
21
Fold: alpha-alpha superhelixSuperfamily: MgtE N-terminal domain-likeFamily: MgtE N-terminal domain-like19 c3k2tA_
26.9
8
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: lmo2511 protein;PDBTitle: crystal structure of lmo2511 protein from listeria2 monocytogenes, northeast structural genomics consortium3 target lkr84a
20 d1xjsa_
26.3
34
Fold: SufE/NifUSuperfamily: SufE/NifUFamily: NifU/IscU domain21 d1fs2b1
not modelled
23.6
33
Fold: Skp1 dimerisation domain-likeSuperfamily: Skp1 dimerisation domain-likeFamily: Skp1 dimerisation domain-like22 d1nexa1
not modelled
19.6
33
Fold: Skp1 dimerisation domain-likeSuperfamily: Skp1 dimerisation domain-likeFamily: Skp1 dimerisation domain-like23 d1dwka1
not modelled
19.4
16
Fold: lambda repressor-like DNA-binding domainsSuperfamily: lambda repressor-like DNA-binding domainsFamily: Cyanase N-terminal domain24 c2zqeA_
not modelled
14.6
36
PDB header: dna binding proteinChain: A: PDB Molecule: muts2 protein;PDBTitle: crystal structure of the smr domain of thermus thermophilus muts2
25 d1pyya2
not modelled
13.6
13
Fold: Penicillin-binding protein 2x (pbp-2x), c-terminal domainSuperfamily: Penicillin-binding protein 2x (pbp-2x), c-terminal domainFamily: Penicillin-binding protein 2x (pbp-2x), c-terminal domain26 d1su0b_
not modelled
13.5
25
Fold: SufE/NifUSuperfamily: SufE/NifUFamily: NifU/IscU domain27 d1rp5a2
not modelled
13.1
14
Fold: Penicillin-binding protein 2x (pbp-2x), c-terminal domainSuperfamily: Penicillin-binding protein 2x (pbp-2x), c-terminal domainFamily: Penicillin-binding protein 2x (pbp-2x), c-terminal domain28 c1rh1A_
not modelled
12.9
21
PDB header: antibioticChain: A: PDB Molecule: colicin b;PDBTitle: crystal structure of the cytotoxic bacterial protein2 colicin b at 2.5 a resolution
29 c3ouvA_
not modelled
12.3
21
PDB header: transferaseChain: A: PDB Molecule: serine/threonine protein kinase;PDBTitle: semet derivative of l512m mutant of pasta domain 3 of mycobacterium2 tuberculosis pknb
30 d1r4va_
not modelled
11.9
29
Fold: Histone-foldSuperfamily: Histone-foldFamily: Bacterial histone-fold protein31 c2i88A_
not modelled
11.5
29
PDB header: membrane proteinChain: A: PDB Molecule: colicin-e1;PDBTitle: crystal structure of the channel-forming domain of colicin2 e1
32 d1k25a2
not modelled
10.6
19
Fold: Penicillin-binding protein 2x (pbp-2x), c-terminal domainSuperfamily: Penicillin-binding protein 2x (pbp-2x), c-terminal domainFamily: Penicillin-binding protein 2x (pbp-2x), c-terminal domain33 c2qq4A_
not modelled
9.5
36
PDB header: metal binding proteinChain: A: PDB Molecule: iron-sulfur cluster biosynthesis protein iscu;PDBTitle: crystal structure of iron-sulfur cluster biosynthesis2 protein iscu (ttha1736) from thermus thermophilus hb8
34 d1ciia1
not modelled
9.2
25
Fold: Toxins' membrane translocation domainsSuperfamily: ColicinFamily: Colicin35 c3fewX_
not modelled
9.0
24
PDB header: immune systemChain: X: PDB Molecule: colicin s4;PDBTitle: structure and function of colicin s4, a colicin with a2 duplicated receptor binding domain
36 c1xc0A_
not modelled
8.4
29
PDB header: signaling proteinChain: A: PDB Molecule: pardaxin p-4;PDBTitle: twenty lowest energy structures of pa4 by solution nmr
37 d2ovra1
not modelled
8.1
25
Fold: Skp1 dimerisation domain-likeSuperfamily: Skp1 dimerisation domain-likeFamily: Skp1 dimerisation domain-like38 d1a87a_
not modelled
8.0
25
Fold: Toxins' membrane translocation domainsSuperfamily: ColicinFamily: Colicin39 c1a87A_
not modelled
8.0
25
PDB header: bacteriocinChain: A: PDB Molecule: colicin n;PDBTitle: colicin n
40 c2p1nD_
not modelled
8.0
40
PDB header: signaling proteinChain: D: PDB Molecule: skp1-like protein 1a;PDBTitle: mechanism of auxin perception by the tir1 ubiqutin ligase
41 d1pyya1
not modelled
8.0
7
Fold: Penicillin-binding protein 2x (pbp-2x), c-terminal domainSuperfamily: Penicillin-binding protein 2x (pbp-2x), c-terminal domainFamily: Penicillin-binding protein 2x (pbp-2x), c-terminal domain42 c2guzD_
not modelled
7.2
19
PDB header: chaperone, protein transportChain: D: PDB Molecule: mitochondrial import inner membrane translocasePDBTitle: structure of the tim14-tim16 complex of the mitochondrial2 protein import motor
43 d1k25a1
not modelled
7.2
14
Fold: Penicillin-binding protein 2x (pbp-2x), c-terminal domainSuperfamily: Penicillin-binding protein 2x (pbp-2x), c-terminal domainFamily: Penicillin-binding protein 2x (pbp-2x), c-terminal domain44 d1v95a_
not modelled
7.2
15
Fold: Anticodon-binding domain-likeSuperfamily: Class II aaRS ABD-relatedFamily: Anticodon-binding domain of Class II aaRS45 c2qdoC_
not modelled
6.9
14
PDB header: photosynthesisChain: C: PDB Molecule: nbla protein;PDBTitle: nbla protein from t. vulcanus
46 d1wwia1
not modelled
6.8
35
Fold: Histone-foldSuperfamily: Histone-foldFamily: Bacterial histone-fold protein47 d1jiha2
not modelled
6.3
13
Fold: DNA/RNA polymerasesSuperfamily: DNA/RNA polymerasesFamily: Lesion bypass DNA polymerase (Y-family), catalytic domain48 d1rh1a2
not modelled
5.8
22
Fold: Toxins' membrane translocation domainsSuperfamily: ColicinFamily: Colicin