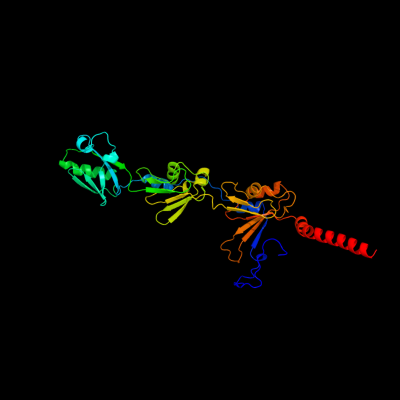
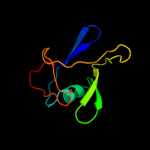
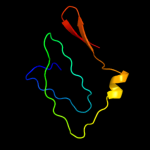
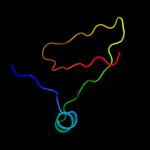
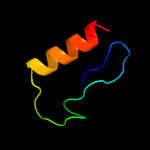
1 c2j58G_
100.0
74
PDB header: membrane proteinChain: G: PDB Molecule: outer membrane lipoprotein wza;PDBTitle: the structure of wza
2 c2w8iG_
100.0
76
PDB header: membrane proteinChain: G: PDB Molecule: putative outer membrane lipoprotein wza;PDBTitle: crystal structure of wza24-345.
3 c3p42D_
99.9
21
PDB header: unknown functionChain: D: PDB Molecule: predicted protein;PDBTitle: structure of gfcc (ymcb), protein encoded by the e. coli group 42 capsule operon
4 d2fug13
88.8
19
Fold: beta-Grasp (ubiquitin-like)Superfamily: Nqo1 middle domain-likeFamily: Nqo1 middle domain-like5 c2fugA_
84.7
17
PDB header: oxidoreductaseChain: A: PDB Molecule: nadh-quinone oxidoreductase chain 1;PDBTitle: crystal structure of the hydrophilic domain of respiratory complex i2 from thermus thermophilus
6 d1ghha_
77.2
18
Fold: DNA damage-inducible protein DinISuperfamily: DNA damage-inducible protein DinIFamily: DNA damage-inducible protein DinI7 c2xdvA_
62.9
16
PDB header: nuclear proteinChain: A: PDB Molecule: myc-induced nuclear antigen;PDBTitle: crystal structure of the catalytic domain of flj14393
8 c3uyjA_
56.8
36
PDB header: oxidoreductaseChain: A: PDB Molecule: lysine-specific demethylase 8;PDBTitle: crystal structure of jmjd5 catalytic core domain in complex with2 nickle and alpha-kg
9 d1vrba1
53.0
19
Fold: Double-stranded beta-helixSuperfamily: Clavaminate synthase-likeFamily: Asparaginyl hydroxylase-like10 d1lr5a_
47.0
14
Fold: Double-stranded beta-helixSuperfamily: RmlC-like cupinsFamily: Germin/Seed storage 7S protein11 c2zpmA_
45.9
29
PDB header: hydrolaseChain: A: PDB Molecule: regulator of sigma e protease;PDBTitle: crystal structure analysis of pdz domain b
12 d1rc6a_
44.6
10
Fold: Double-stranded beta-helixSuperfamily: RmlC-like cupinsFamily: YlbA-like13 c3al6A_
43.4
35
PDB header: unknown functionChain: A: PDB Molecule: jmjc domain-containing protein c2orf60;PDBTitle: crystal structure of human tyw5
14 d1zofa1
43.3
14
Fold: Thioredoxin foldSuperfamily: Thioredoxin-likeFamily: Glutathione peroxidase-like15 c3u7zA_
41.0
11
PDB header: metal binding proteinChain: A: PDB Molecule: putative metal binding protein rumgna_00854;PDBTitle: crystal structure of a putative metal binding protein rumgna_008542 (zp_02040092.1) from ruminococcus gnavus atcc 29149 at 1.30 a3 resolution
16 d1x82a_
39.8
8
Fold: Double-stranded beta-helixSuperfamily: RmlC-like cupinsFamily: Glucose-6-phosphate isomerase, GPI17 c2xxzA_
39.8
19
PDB header: oxidoreductaseChain: A: PDB Molecule: lysine-specific demethylase 6b;PDBTitle: crystal structure of the human jmjd3 jumonji domain
18 c3eytA_
39.2
16
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: uncharacterized protein spoa0173;PDBTitle: crystal structure of thioredoxin-like superfamily protein spoa0173
19 c2opkC_
38.1
16
PDB header: isomeraseChain: C: PDB Molecule: hypothetical protein;PDBTitle: crystal structure of a putative mannose-6-phosphate isomerase2 (reut_a1446) from ralstonia eutropha jmp134 at 2.10 a resolution
20 c3k2oB_
37.8
38
PDB header: oxidoreductaseChain: B: PDB Molecule: bifunctional arginine demethylase and lysyl-hydroxylasePDBTitle: structure of an oxygenase
21 c3razA_
not modelled
37.3
3
PDB header: oxidoreductaseChain: A: PDB Molecule: thioredoxin-related protein;PDBTitle: the crystal structure of thioredoxin-related protein from neisseria2 meningitidis serogroup b
22 d1fe0a_
not modelled
37.1
14
Fold: Ferredoxin-likeSuperfamily: HMA, heavy metal-associated domainFamily: HMA, heavy metal-associated domain23 d1h2ka_
not modelled
35.3
29
Fold: Double-stranded beta-helixSuperfamily: Clavaminate synthase-likeFamily: Hypoxia-inducible factor HIF ihhibitor (FIH1)24 d1wp0a1
not modelled
33.2
12
Fold: Thioredoxin foldSuperfamily: Thioredoxin-likeFamily: Glutathione peroxidase-like25 c3lorB_
not modelled
32.5
16
PDB header: isomeraseChain: B: PDB Molecule: thiol-disulfide isomerase and thioredoxins;PDBTitle: the crystal structure of a thiol-disulfide isomerase from2 corynebacterium glutamicum to 2.2a
26 d1sfna_
not modelled
32.4
15
Fold: Double-stranded beta-helixSuperfamily: RmlC-like cupinsFamily: YlbA-like27 d1rwua_
not modelled
32.1
12
Fold: Ferredoxin-likeSuperfamily: YbeD/HP0495-likeFamily: YbeD-like28 c1rwuA_
not modelled
32.1
12
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: hypothetical upf0250 protein ybed;PDBTitle: solution structure of conserved protein ybed from e. coli
29 c3cewA_
not modelled
31.9
21
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: uncharacterized cupin protein;PDBTitle: crystal structure of a cupin protein (bf4112) from bacteroides2 fragilis. northeast structural genomics consortium target bfr205
30 c2yztA_
not modelled
31.5
6
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: putative uncharacterized protein ttha1756;PDBTitle: crystal structure of uncharacterized conserved protein from thermus2 thermophilus hb8
31 d1zx5a1
not modelled
31.1
24
Fold: Double-stranded beta-helixSuperfamily: RmlC-like cupinsFamily: Type I phosphomannose isomerase32 d1sq4a_
not modelled
31.0
9
Fold: Double-stranded beta-helixSuperfamily: RmlC-like cupinsFamily: YlbA-like33 c1sefA_
not modelled
30.9
11
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: conserved hypothetical protein;PDBTitle: crystal structure of cupin domain protein ef2996 from enterococcus2 faecalis
34 d1sefa_
not modelled
30.9
11
Fold: Double-stranded beta-helixSuperfamily: RmlC-like cupinsFamily: YlbA-like35 c2r37A_
not modelled
30.8
22
PDB header: oxidoreductaseChain: A: PDB Molecule: glutathione peroxidase 3;PDBTitle: crystal structure of human glutathione peroxidase 3 (selenocysteine to2 glycine mutant)
36 c2r32A_
not modelled
29.3
18
PDB header: immune systemChain: A: PDB Molecule: gcn4-pii/tumor necrosis factor ligandPDBTitle: crystal structure of human gitrl variant
37 d1z5ye1
not modelled
27.7
17
Fold: Thioredoxin foldSuperfamily: Thioredoxin-likeFamily: Glutathione peroxidase-like38 d1y7ma2
not modelled
27.6
4
Fold: LysM domainSuperfamily: LysM domainFamily: LysM domain39 c3ibmB_
not modelled
27.2
21
PDB header: structural genomics, unknown functionChain: B: PDB Molecule: cupin 2, conserved barrel domain protein;PDBTitle: crystal structure of cupin 2 domain-containing protein hhal_0468 from2 halorhodospira halophila
40 d1we0a1
not modelled
27.0
23
Fold: Thioredoxin foldSuperfamily: Thioredoxin-likeFamily: Glutathione peroxidase-like41 d1q44a_
not modelled
26.6
17
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: PAPS sulfotransferase42 c3d0fA_
not modelled
26.4
18
PDB header: transferaseChain: A: PDB Molecule: penicillin-binding 1 transmembrane protein mrca;PDBTitle: structure of the big_1156.2 domain of putative penicillin-binding2 protein mrca from nitrosomonas europaea atcc 19718
43 d1qmva_
not modelled
26.3
13
Fold: Thioredoxin foldSuperfamily: Thioredoxin-likeFamily: Glutathione peroxidase-like44 d1j3pa_
not modelled
26.0
10
Fold: Double-stranded beta-helixSuperfamily: RmlC-like cupinsFamily: Glucose-6-phosphate isomerase, GPI45 c2h8kA_
not modelled
25.3
14
PDB header: transferaseChain: A: PDB Molecule: sult1c3 splice variant d;PDBTitle: human sulfotranferase sult1c3 in complex with pap
46 c3cmiA_
not modelled
25.0
19
PDB header: oxidoreductaseChain: A: PDB Molecule: peroxiredoxin hyr1;PDBTitle: crystal structure of glutathione-dependent phospholipid peroxidase2 hyr1 from the yeast saccharomyces cerevisiae
47 d2dsya1
not modelled
24.6
20
Fold: TTHA1013/TTHA0281-likeSuperfamily: TTHA1013/TTHA0281-likeFamily: TTHA0281-like48 c3kh7A_
not modelled
24.6
15
PDB header: oxidoreductaseChain: A: PDB Molecule: thiol:disulfide interchange protein dsbe;PDBTitle: crystal structure of the periplasmic soluble domain of reduced ccmg2 from pseudomonas aeruginosa
49 c3d82A_
not modelled
24.3
30
PDB header: metal binding proteinChain: A: PDB Molecule: cupin 2, conserved barrel domain protein;PDBTitle: crystal structure of a cupin-2 domain containing protein (sfri_3543)2 from shewanella frigidimarina ncimb 400 at 2.05 a resolution
50 d1knga_
not modelled
23.9
22
Fold: Thioredoxin foldSuperfamily: Thioredoxin-likeFamily: Glutathione peroxidase-like51 d1uija2
not modelled
23.6
8
Fold: Double-stranded beta-helixSuperfamily: RmlC-like cupinsFamily: Germin/Seed storage 7S protein52 c2v1mA_
not modelled
23.4
16
PDB header: oxidoreductaseChain: A: PDB Molecule: glutathione peroxidase;PDBTitle: crystal structure of schistosoma mansoni glutathione2 peroxidase
53 c3or5A_
not modelled
22.7
13
PDB header: oxidoreductaseChain: A: PDB Molecule: thiol:disulfide interchange protein, thioredoxin familyPDBTitle: crystal structure of thiol:disulfide interchange protein, thioredoxin2 family protein from chlorobium tepidum tls
54 c2rogA_
not modelled
22.7
30
PDB header: metal binding proteinChain: A: PDB Molecule: heavy metal binding protein;PDBTitle: solution structure of thermus thermophilus hb8 ttha17182 protein in living e. coli cells
55 c3h8uA_
not modelled
22.5
18
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: uncharacterized conserved protein with double-strandedPDBTitle: crystal structure of uncharacterized conserved protein with double-2 stranded beta-helix domain (yp_001338853.1) from klebsiella3 pneumoniae subsp. pneumoniae mgh 78578 at 1.80 a resolution
56 d1fmja_
not modelled
22.5
14
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: PAPS sulfotransferase57 c2hhzA_
not modelled
22.3
11
PDB header: oxidoreductaseChain: A: PDB Molecule: pyridoxamine 5'-phosphate oxidase-related;PDBTitle: crystal structure of a pyridoxamine 5'-phosphate oxidase-related2 protein (ssuidraft_2804) from streptococcus suis 89/1591 at 2.00 a3 resolution
58 c3lwaA_
not modelled
22.3
9
PDB header: isomeraseChain: A: PDB Molecule: secreted thiol-disulfide isomerase;PDBTitle: the crystal structure of a secreted thiol-disulfide2 isomerase from corynebacterium glutamicum to 1.75a
59 c3jxoB_
not modelled
22.3
11
PDB header: transport proteinChain: B: PDB Molecule: trka-n domain protein;PDBTitle: crystal structure of an octomeric two-subunit trka k+ channel ring2 gating assembly, tm1088a:tm1088b, from thermotoga maritima
60 c3drnB_
not modelled
22.3
8
PDB header: oxidoreductaseChain: B: PDB Molecule: peroxiredoxin, bacterioferritin comigratory proteinPDBTitle: the crystal structure of bcp1 from sulfolobus sulfataricus
61 d1prxa_
not modelled
22.1
4
Fold: Thioredoxin foldSuperfamily: Thioredoxin-likeFamily: Glutathione peroxidase-like62 d1gp1a_
not modelled
22.0
20
Fold: Thioredoxin foldSuperfamily: Thioredoxin-likeFamily: Glutathione peroxidase-like63 c3h7hA_
not modelled
22.0
21
PDB header: transcriptionChain: A: PDB Molecule: transcription elongation factor spt4;PDBTitle: crystal structure of the human transcription elongation factor dsif,2 hspt4/hspt5 (176-273)
64 d1uika2
not modelled
21.9
11
Fold: Double-stranded beta-helixSuperfamily: RmlC-like cupinsFamily: Germin/Seed storage 7S protein65 d1te7a_
not modelled
21.3
20
Fold: PUA domain-likeSuperfamily: PUA domain-likeFamily: yqfB-like66 d2d40a1
not modelled
21.3
13
Fold: Double-stranded beta-helixSuperfamily: RmlC-like cupinsFamily: Gentisate 1,2-dioxygenase-like67 c2wbqA_
not modelled
21.1
15
PDB header: oxidoreductaseChain: A: PDB Molecule: l-arginine beta-hydroxylase;PDBTitle: crystal structure of vioc in complex with (2s,3s)-2 hydroxyarginine
68 d1pmia_
not modelled
20.4
7
Fold: Double-stranded beta-helixSuperfamily: RmlC-like cupinsFamily: Type I phosphomannose isomerase69 c2q30C_
not modelled
20.4
17
PDB header: unknown functionChain: C: PDB Molecule: uncharacterized protein;PDBTitle: crystal structure of a rmlc-like cupin protein (dde_2303) from2 desulfovibrio desulfuricans subsp. at 1.94 a resolution
70 c3h1yA_
not modelled
20.1
15
PDB header: isomeraseChain: A: PDB Molecule: mannose-6-phosphate isomerase;PDBTitle: crystal structure of mannose 6-phosphate isomerase from2 salmonella typhimurium bound to substrate (f6p)and metal3 atom (zn)
71 d1xv1a_
not modelled
20.1
21
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: PAPS sulfotransferase72 d2phna1
not modelled
19.8
14
Fold: CofE-likeSuperfamily: CofE-likeFamily: CofE-like73 d1q20a_
not modelled
19.4
14
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: PAPS sulfotransferase74 c2h66G_
not modelled
19.3
12
PDB header: structural genomics/oxidoreductaseChain: G: PDB Molecule: pv-pf14_0368;PDBTitle: the crystal structure of plasmodium vivax 2-cys2 peroxiredoxin
75 d1yexa1
not modelled
19.2
18
Fold: Thioredoxin foldSuperfamily: Thioredoxin-likeFamily: Glutathione peroxidase-like76 c2og5A_
not modelled
19.1
23
PDB header: oxidoreductaseChain: A: PDB Molecule: putative oxygenase;PDBTitle: crystal structure of asparagine oxygenase (asno)
77 d1e2ya_
not modelled
18.9
15
Fold: Thioredoxin foldSuperfamily: Thioredoxin-likeFamily: Glutathione peroxidase-like78 d1ds1a_
not modelled
18.7
38
Fold: Double-stranded beta-helixSuperfamily: Clavaminate synthase-likeFamily: Clavaminate synthase79 d2b8ma1
not modelled
18.7
14
Fold: Double-stranded beta-helixSuperfamily: RmlC-like cupinsFamily: MJ0764-like80 c3bcwB_
not modelled
18.5
23
PDB header: unknown functionChain: B: PDB Molecule: uncharacterized protein;PDBTitle: crystal structure of a duf861 family protein with a rmlc-like cupin2 fold (bb1179) from bordetella bronchiseptica rb50 at 1.60 a3 resolution
81 d1o5ua_
not modelled
18.1
27
Fold: Double-stranded beta-helixSuperfamily: RmlC-like cupinsFamily: Hypothetical protein TM111282 c3kw0D_
not modelled
18.0
17
PDB header: hydrolaseChain: D: PDB Molecule: cysteine peptidase;PDBTitle: crystal structure of cysteine peptidase (np_982244.1) from bacillus2 cereus atcc 10987 at 2.50 a resolution
83 d1j99a_
not modelled
18.0
14
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: PAPS sulfotransferase84 c3ia1A_
not modelled
17.8
13
PDB header: oxidoreductaseChain: A: PDB Molecule: thio-disulfide isomerase/thioredoxin;PDBTitle: crystal structure of thio-disulfide isomerase from thermus2 thermophilus
85 c2v2gC_
not modelled
17.7
9
PDB header: oxidoreductaseChain: C: PDB Molecule: peroxiredoxin 6;PDBTitle: crystal structure of the c45s mutant of the peroxiredoxin 62 of arenicola marina. monoclinic form
86 c2hc8A_
not modelled
17.5
21
PDB header: transport proteinChain: A: PDB Molecule: cation-transporting atpase, p-type;PDBTitle: structure of the a. fulgidus copa a-domain
87 d1sb6a_
not modelled
17.4
21
Fold: Ferredoxin-likeSuperfamily: HMA, heavy metal-associated domainFamily: HMA, heavy metal-associated domain88 c3jzvA_
not modelled
17.4
17
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: uncharacterized protein rru_a2000;PDBTitle: crystal structure of rru_a2000 from rhodospirillum rubrum: a cupin-22 domain.
89 c3gl3D_
not modelled
16.9
16
PDB header: oxidoreductaseChain: D: PDB Molecule: putative thiol:disulfide interchange proteinPDBTitle: crystal structure of a putative thiol:disulfide interchange2 protein dsbe from chlorobium tepidum
90 d1zud21
not modelled
16.9
7
Fold: beta-Grasp (ubiquitin-like)Superfamily: MoaD/ThiSFamily: ThiS91 d1qq2a_
not modelled
16.9
16
Fold: Thioredoxin foldSuperfamily: Thioredoxin-likeFamily: Glutathione peroxidase-like92 c2fpeB_
not modelled
16.9
20
PDB header: signaling proteinChain: B: PDB Molecule: c-jun-amino-terminal kinase interacting proteinPDBTitle: conserved dimerization of the ib1 src-homology 3 domain
93 c2rqeA_
not modelled
16.7
17
PDB header: sugar binding proteinChain: A: PDB Molecule: beta-1,3-glucan-binding protein;PDBTitle: solution structure of the silkworm bgrp/gnbp3 n-terminal2 domain reveals the mechanism for b-1,3-glucan specific3 recognition
94 c3k6qB_
not modelled
16.1
15
PDB header: ligand binding proteinChain: B: PDB Molecule: putative ligand binding protein;PDBTitle: crystal structure of an antitoxin part of a putative toxin/antitoxin2 system (swol_0700) from syntrophomonas wolfei subsp. wolfei at 1.80 a3 resolution
95 c2kytA_
not modelled
16.1
14
PDB header: hydrolaseChain: A: PDB Molecule: group xvi phospholipase a2;PDBTitle: solution struture of the h-rev107 n-terminal domain
96 d1qwra_
not modelled
16.0
17
Fold: Double-stranded beta-helixSuperfamily: RmlC-like cupinsFamily: Type I phosphomannose isomerase97 d1g3ma_
not modelled
15.4
17
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: PAPS sulfotransferase98 c2gu9B_
not modelled
15.4
20
PDB header: immune systemChain: B: PDB Molecule: tetracenomycin polyketide synthesis protein;PDBTitle: crystal structure of xc5357 from xanthomonas campestris: a2 putative tetracenomycin polyketide synthesis protein3 adopting a novel cupin subfamily structure
99 d1xcca_
not modelled
15.4
9
Fold: Thioredoxin foldSuperfamily: Thioredoxin-likeFamily: Glutathione peroxidase-like