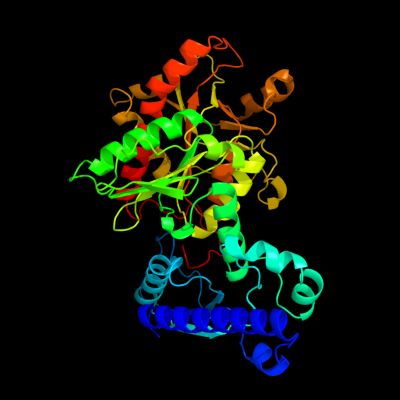
| 1 | d1t8sa_
|
|
 |
100.0 |
99 |
Fold:Phosphorylase/hydrolase-like
Superfamily:Purine and uridine phosphorylases
Family:Purine and uridine phosphorylases |
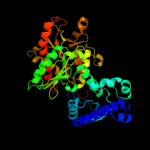
| 2 | d1ybfa_
|
|
 |
100.0 |
32 |
Fold:Phosphorylase/hydrolase-like
Superfamily:Purine and uridine phosphorylases
Family:Purine and uridine phosphorylases |
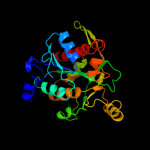
| 3 | c3mb8A_
|
|
 |
100.0 |
19 |
PDB header:transferase
Chain: A: PDB Molecule:purine nucleoside phosphorylase;
PDBTitle: crystal structure of purine nucleoside phosphorylase from toxoplasma2 gondii in complex with immucillin-h
|
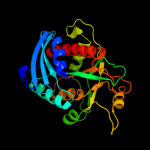
| 4 | c1nw4C_
|
|
 |
100.0 |
22 |
PDB header:transferase
Chain: C: PDB Molecule:uridine phosphorylase, putative;
PDBTitle: crystal structure of plasmodium falciparum purine nucleoside2 phosphorylase in complex with immh and sulfate
|
| 5 | d1q1ga_
|
|
 |
100.0 |
22 |
Fold:Phosphorylase/hydrolase-like
Superfamily:Purine and uridine phosphorylases
Family:Purine and uridine phosphorylases |
| 6 | c3qpbB_
|
|
 |
100.0 |
22 |
PDB header:transferase
Chain: B: PDB Molecule:uridine phosphorylase;
PDBTitle: crystal structure of streptococcus pyogenes uridine phosphorylase2 reveals a subclass of the np-i superfamily
|
| 7 | d1odka_
|
|
 |
100.0 |
20 |
Fold:Phosphorylase/hydrolase-like
Superfamily:Purine and uridine phosphorylases
Family:Purine and uridine phosphorylases |
| 8 | c3bjeA_
|
|
 |
100.0 |
18 |
PDB header:transferase
Chain: A: PDB Molecule:nucleoside phosphorylase, putative;
PDBTitle: crystal structure of trypanosoma brucei nucleoside phosphorylase shows2 uridine phosphorylase activity
|
| 9 | d1k9sa_
|
|
 |
100.0 |
17 |
Fold:Phosphorylase/hydrolase-like
Superfamily:Purine and uridine phosphorylases
Family:Purine and uridine phosphorylases |
| 10 | d1vhwa_
|
|
 |
100.0 |
16 |
Fold:Phosphorylase/hydrolase-like
Superfamily:Purine and uridine phosphorylases
Family:Purine and uridine phosphorylases |
| 11 | d1rxya_
|
|
 |
100.0 |
21 |
Fold:Phosphorylase/hydrolase-like
Superfamily:Purine and uridine phosphorylases
Family:Purine and uridine phosphorylases |
| 12 | c1z34A_
|
|
 |
100.0 |
22 |
PDB header:transferase
Chain: A: PDB Molecule:purine nucleoside phosphorylase;
PDBTitle: crystal structure of trichomonas vaginalis purine nucleoside2 phosphorylase complexed with 2-fluoro-2'-deoxyadenosine
|
| 13 | c2xrfA_
|
|
 |
100.0 |
16 |
PDB header:transferase
Chain: A: PDB Molecule:uridine phosphorylase 2;
PDBTitle: crystal structure of human uridine phosphorylase 2
|
| 14 | d1je0a_
|
|
 |
100.0 |
18 |
Fold:Phosphorylase/hydrolase-like
Superfamily:Purine and uridine phosphorylases
Family:Purine and uridine phosphorylases |
| 15 | c3tl6B_
|
|
 |
100.0 |
17 |
PDB header:transferase
Chain: B: PDB Molecule:purine nucleoside phosphorylase;
PDBTitle: crystal structure of purine nucleoside phosphorylase from entamoeba2 histolytica
|
| 16 | c3eufC_
|
|
 |
100.0 |
16 |
PDB header:transferase
Chain: C: PDB Molecule:uridine phosphorylase 1;
PDBTitle: crystal structure of bau-bound human uridine phosphorylase 1
|
| 17 | d2ac7a1
|
|
 |
100.0 |
22 |
Fold:Phosphorylase/hydrolase-like
Superfamily:Purine and uridine phosphorylases
Family:Purine and uridine phosphorylases |
| 18 | c3bl6A_
|
|
 |
100.0 |
20 |
PDB header:hydrolase
Chain: A: PDB Molecule:5'-methylthioadenosine nucleosidase/s-
PDBTitle: crystal structure of staphylococcus aureus 5'-2 methylthioadenosine/s-adenosylhomocysteine nucleosidase in3 complex with formycin a
|
| 19 | c3eeiA_
|
|
 |
100.0 |
20 |
PDB header:hydrolase
Chain: A: PDB Molecule:5-methylthioadenosine nucleosidase/s-
PDBTitle: crystal structure of 5'-methylthioadenosine/s-2 adenosylhomocysteine nucleosidase from neisseria3 meningitidis in complex with methylthio-immucillin-a
|
| 20 | c3dp9A_
|
|
 |
100.0 |
18 |
PDB header:hydrolase
Chain: A: PDB Molecule:mta/sah nucleosidase;
PDBTitle: crystal structure of vibrio cholerae 5'-methylthioadenosine/s-adenosyl2 homocysteine nucleosidase (mtan) complexed with butylthio-dadme-3 immucillin a
|
| 21 | c3nm5B_ |
|
not modelled |
100.0 |
18 |
PDB header:hydrolase
Chain: B: PDB Molecule:mta/sah nucleosidase;
PDBTitle: helicobacter pylori mtan complexed with formycin a
|
| 22 | c1zosE_ |
|
not modelled |
100.0 |
20 |
PDB header:hydrolase
Chain: E: PDB Molecule:5'-methylthioadenosine / s-adenosylhomocysteine
PDBTitle: structure of 5'-methylthionadenosine/s-adenosylhomocysteine2 nucleosidase from s. pneumoniae with a transition-state3 inhibitor mt-imma
|
| 23 | d1jysa_ |
|
not modelled |
100.0 |
22 |
Fold:Phosphorylase/hydrolase-like
Superfamily:Purine and uridine phosphorylases
Family:Purine and uridine phosphorylases |
| 24 | c2h8gA_ |
|
not modelled |
100.0 |
20 |
PDB header:hydrolase
Chain: A: PDB Molecule:5'-methylthioadenosine nucleosidase;
PDBTitle: 5'-methylthioadenosine nucleosidase from arabidopsis2 thaliana
|
| 25 | c3bsfB_ |
|
not modelled |
100.0 |
19 |
PDB header:hydrolase
Chain: B: PDB Molecule:at4g34840;
PDBTitle: crystal structure of the mta/sah nucleosidase
|
| 26 | c3ozbF_ |
|
not modelled |
99.9 |
13 |
PDB header:transferase
Chain: F: PDB Molecule:methylthioadenosine phosphorylase;
PDBTitle: crystal structure of 5'-methylthioinosine phosphorylase from2 psedomonas aeruginosa in complex with hypoxanthine
|
| 27 | c1wtaA_ |
|
not modelled |
99.9 |
15 |
PDB header:transferase
Chain: A: PDB Molecule:5'-methylthioadenosine phosphorylase;
PDBTitle: crystal structure of 5'-deoxy-5'-methylthioadenosine from aeropyrum2 pernix (r32 form)
|
| 28 | d1g2oa_ |
|
not modelled |
99.9 |
12 |
Fold:Phosphorylase/hydrolase-like
Superfamily:Purine and uridine phosphorylases
Family:Purine and uridine phosphorylases |
| 29 | d1v4na_ |
|
not modelled |
99.9 |
13 |
Fold:Phosphorylase/hydrolase-like
Superfamily:Purine and uridine phosphorylases
Family:Purine and uridine phosphorylases |
| 30 | c3khsB_ |
|
not modelled |
99.9 |
15 |
PDB header:hydrolase
Chain: B: PDB Molecule:purine nucleoside phosphorylase;
PDBTitle: crystal structure of grouper iridovirus purine nucleoside2 phosphorylase
|
| 31 | d1vmka_ |
|
not modelled |
99.8 |
9 |
Fold:Phosphorylase/hydrolase-like
Superfamily:Purine and uridine phosphorylases
Family:Purine and uridine phosphorylases |
| 32 | c2p4sA_ |
|
not modelled |
99.8 |
12 |
PDB header:transferase
Chain: A: PDB Molecule:purine nucleoside phosphorylase;
PDBTitle: structure of purine nucleoside phosphorylase from anopheles gambiae in2 complex with dadme-immh
|
| 33 | d1cb0a_ |
|
not modelled |
99.8 |
13 |
Fold:Phosphorylase/hydrolase-like
Superfamily:Purine and uridine phosphorylases
Family:Purine and uridine phosphorylases |
| 34 | c1yr3A_ |
|
not modelled |
99.8 |
13 |
PDB header:transferase
Chain: A: PDB Molecule:xanthosine phosphorylase;
PDBTitle: escherichia coli purine nucleoside phosphorylase ii, the2 product of the xapa gene
|
| 35 | d1qe5a_ |
|
not modelled |
99.8 |
15 |
Fold:Phosphorylase/hydrolase-like
Superfamily:Purine and uridine phosphorylases
Family:Purine and uridine phosphorylases |
| 36 | c1tcvB_ |
|
not modelled |
99.8 |
10 |
PDB header:transferase
Chain: B: PDB Molecule:purine-nucleoside phosphorylase;
PDBTitle: crystal structure of the purine nucleoside phosphorylase2 from schistosoma mansoni in complex with non-detergent3 sulfobetaine 195 and acetate
|
| 37 | c3la8A_ |
|
not modelled |
99.8 |
10 |
PDB header:transferase
Chain: A: PDB Molecule:putative purine nucleoside phosphorylase;
PDBTitle: the crystal structure of smu.1229 from streptococcus mutans ua159
|
| 38 | d3bgsa1 |
|
not modelled |
99.8 |
15 |
Fold:Phosphorylase/hydrolase-like
Superfamily:Purine and uridine phosphorylases
Family:Purine and uridine phosphorylases |
| 39 | d3pnpa_ |
|
not modelled |
99.8 |
13 |
Fold:Phosphorylase/hydrolase-like
Superfamily:Purine and uridine phosphorylases
Family:Purine and uridine phosphorylases |
| 40 | c3ggsA_ |
|
not modelled |
99.7 |
12 |
PDB header:transferase
Chain: A: PDB Molecule:purine nucleoside phosphorylase;
PDBTitle: human purine nucleoside phosphorylase double mutant e201q,n243d2 complexed with 2-fluoro-2'-deoxyadenosine
|
| 41 | d1ixka_ |
|
not modelled |
74.3 |
14 |
Fold:S-adenosyl-L-methionine-dependent methyltransferases
Superfamily:S-adenosyl-L-methionine-dependent methyltransferases
Family:NOL1/NOP2/sun |
| 42 | d1sv6a_ |
|
not modelled |
66.4 |
20 |
Fold:FAH
Superfamily:FAH
Family:FAH |
| 43 | c3lywA_ |
|
not modelled |
60.9 |
19 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:ybbr family protein;
PDBTitle: crystal structure of ybbr family protein dhaf_0833 from2 desulfitobacterium hafniense dcb-2. northeast structural3 genomics consortium target id dhr29b
|
| 44 | c3m4xA_ |
|
not modelled |
53.7 |
11 |
PDB header:transferase
Chain: A: PDB Molecule:nol1/nop2/sun family protein;
PDBTitle: structure of a ribosomal methyltransferase
|
| 45 | c2yxlA_ |
|
not modelled |
48.7 |
16 |
PDB header:transferase
Chain: A: PDB Molecule:450aa long hypothetical fmu protein;
PDBTitle: crystal structure of ph0851
|
| 46 | c2eb5D_ |
|
not modelled |
44.0 |
14 |
PDB header:lyase
Chain: D: PDB Molecule:2-oxo-hept-3-ene-1,7-dioate hydratase;
PDBTitle: crystal structure of hpcg complexed with oxalate
|
| 47 | d1nr9a_ |
|
not modelled |
42.9 |
14 |
Fold:FAH
Superfamily:FAH
Family:FAH |
| 48 | d1sgva2 |
|
not modelled |
41.2 |
23 |
Fold:Pseudouridine synthase
Superfamily:Pseudouridine synthase
Family:Pseudouridine synthase II TruB |
| 49 | d1gtta2 |
|
not modelled |
40.8 |
18 |
Fold:FAH
Superfamily:FAH
Family:FAH |
| 50 | d2es9a1 |
|
not modelled |
37.9 |
40 |
Fold:YoaC-like
Superfamily:YoaC-like
Family:YoaC-like |
| 51 | d1gpua3 |
|
not modelled |
37.6 |
16 |
Fold:TK C-terminal domain-like
Superfamily:TK C-terminal domain-like
Family:Transketolase C-terminal domain-like |
| 52 | c3r6oA_ |
|
not modelled |
37.1 |
18 |
PDB header:isomerase
Chain: A: PDB Molecule:2-hydroxyhepta-2,4-diene-1, 7-dioateisomerase;
PDBTitle: crystal structure of a probable 2-hydroxyhepta-2,4-diene-1, 7-2 dioateisomerase from mycobacterium abscessus
|
| 53 | c2dfuB_ |
|
not modelled |
36.7 |
13 |
PDB header:isomerase
Chain: B: PDB Molecule:probable 2-hydroxyhepta-2,4-diene-1,7-dioate isomerase;
PDBTitle: crystal structure of the 2-hydroxyhepta-2,4-diene-1,7-dioate isomerase2 from thermus thermophilus hb8
|
| 54 | c1ojhK_ |
|
not modelled |
36.6 |
47 |
PDB header:protein binding
Chain: K: PDB Molecule:nbla;
PDBTitle: crystal structure of nbla from pcc 7120
|
| 55 | d1ojha_ |
|
not modelled |
35.0 |
47 |
Fold:NblA-like
Superfamily:NblA-like
Family:NblA-like |
| 56 | c1sqgA_ |
|
not modelled |
34.6 |
18 |
PDB header:transferase
Chain: A: PDB Molecule:sun protein;
PDBTitle: the crystal structure of the e. coli fmu apoenzyme at 1.652 a resolution
|
| 57 | d1gtta1 |
|
not modelled |
33.8 |
21 |
Fold:FAH
Superfamily:FAH
Family:FAH |
| 58 | c3g7gG_ |
|
not modelled |
32.5 |
19 |
PDB header:structural genomics, unknown function
Chain: G: PDB Molecule:upf0311 protein ca_c3321;
PDBTitle: crystal structure of the protein with unknown function from2 clostridium acetobutylicum atcc 824
|
| 59 | d1nkqa_ |
|
not modelled |
31.1 |
29 |
Fold:FAH
Superfamily:FAH
Family:FAH |
| 60 | d1qcza_ |
|
not modelled |
30.1 |
20 |
Fold:Flavodoxin-like
Superfamily:N5-CAIR mutase (phosphoribosylaminoimidazole carboxylase, PurE)
Family:N5-CAIR mutase (phosphoribosylaminoimidazole carboxylase, PurE) |
| 61 | c1a4iB_ |
|
not modelled |
30.0 |
16 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:methylenetetrahydrofolate dehydrogenase /
PDBTitle: human tetrahydrofolate dehydrogenase / cyclohydrolase
|
| 62 | d2cl5a1 |
|
not modelled |
26.5 |
21 |
Fold:S-adenosyl-L-methionine-dependent methyltransferases
Superfamily:S-adenosyl-L-methionine-dependent methyltransferases
Family:COMT-like |
| 63 | d1b0aa1 |
|
not modelled |
25.8 |
14 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Aminoacid dehydrogenase-like, C-terminal domain |
| 64 | c3qdfA_ |
|
not modelled |
25.3 |
18 |
PDB header:isomerase
Chain: A: PDB Molecule:2-hydroxyhepta-2,4-diene-1,7-dioate isomerase;
PDBTitle: crystal structure of 2-hydroxyhepta-2,4-diene-1,7-dioate isomerase2 from mycobacterium marinum
|
| 65 | c3c5oD_ |
|
not modelled |
25.1 |
17 |
PDB header:structural genomics, unknown function
Chain: D: PDB Molecule:upf0311 protein rpa1785;
PDBTitle: crystal structure of the conserved protein of unknown function rpa17852 from rhodopseudomonas palustris
|
| 66 | c1wzoC_ |
|
not modelled |
24.7 |
14 |
PDB header:isomerase
Chain: C: PDB Molecule:hpce;
PDBTitle: crystal structure of the hpce from thermus thermophilus hb8
|
| 67 | c3p2oA_ |
|
not modelled |
23.7 |
16 |
PDB header:oxidoreductase, hydrolase
Chain: A: PDB Molecule:bifunctional protein fold;
PDBTitle: crystal structure of fold bifunctional protein from campylobacter2 jejuni
|
| 68 | d1sawa_ |
|
not modelled |
22.9 |
13 |
Fold:FAH
Superfamily:FAH
Family:FAH |
| 69 | d1a4ia1 |
|
not modelled |
22.8 |
20 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Aminoacid dehydrogenase-like, C-terminal domain |
| 70 | d1p9ea_ |
|
not modelled |
22.4 |
24 |
Fold:Metallo-hydrolase/oxidoreductase
Superfamily:Metallo-hydrolase/oxidoreductase
Family:Methyl parathion hydrolase |
| 71 | c1p9eA_ |
|
not modelled |
22.4 |
24 |
PDB header:hydrolase
Chain: A: PDB Molecule:methyl parathion hydrolase;
PDBTitle: crystal structure analysis of methyl parathion hydrolase from2 pseudomonas sp wbc-3
|
| 72 | c2h31A_ |
|
not modelled |
21.9 |
15 |
PDB header:ligase, lyase
Chain: A: PDB Molecule:multifunctional protein ade2;
PDBTitle: crystal structure of human paics, a bifunctional carboxylase and2 synthetase in purine biosynthesis
|
| 73 | d1sqga2 |
|
not modelled |
21.9 |
17 |
Fold:S-adenosyl-L-methionine-dependent methyltransferases
Superfamily:S-adenosyl-L-methionine-dependent methyltransferases
Family:NOL1/NOP2/sun |
| 74 | d1o4va_ |
|
not modelled |
21.6 |
22 |
Fold:Flavodoxin-like
Superfamily:N5-CAIR mutase (phosphoribosylaminoimidazole carboxylase, PurE)
Family:N5-CAIR mutase (phosphoribosylaminoimidazole carboxylase, PurE) |
| 75 | c3p2oB_ |
|
not modelled |
21.5 |
16 |
PDB header:oxidoreductase, hydrolase
Chain: B: PDB Molecule:bifunctional protein fold;
PDBTitle: crystal structure of fold bifunctional protein from campylobacter2 jejuni
|
| 76 | c3eshB_ |
|
not modelled |
21.2 |
27 |
PDB header:hydrolase
Chain: B: PDB Molecule:protein similar to metal-dependent hydrolase;
PDBTitle: crystal structure of a probable metal-dependent hydrolase2 from staphylococcus aureus. northeast structural genomics3 target zr314
|
| 77 | c3m6wA_ |
|
not modelled |
19.7 |
17 |
PDB header:transferase
Chain: A: PDB Molecule:rrna methylase;
PDBTitle: multi-site-specific 16s rrna methyltransferase rsmf from thermus2 thermophilus in space group p21212 in complex with s-adenosyl-l-3 methionine
|
| 78 | c2q1dX_ |
|
not modelled |
18.3 |
13 |
PDB header:lyase
Chain: X: PDB Molecule:2-keto-3-deoxy-d-arabinonate dehydratase;
PDBTitle: 2-keto-3-deoxy-d-arabinonate dehydratase complexed with magnesium and2 2,5-dioxopentanoate
|
| 79 | c2k5jB_ |
|
not modelled |
17.8 |
20 |
PDB header:structural genomics, unknown function
Chain: B: PDB Molecule:uncharacterized protein yiif;
PDBTitle: solution structure of protein yiif from shigella flexneri2 serotype 5b (strain 8401) . northeast structural genomics3 consortium target sft1
|
| 80 | c3l53F_ |
|
not modelled |
17.6 |
18 |
PDB header:structural genomics, unknown function
Chain: F: PDB Molecule:putative fumarylacetoacetate isomerase/hydrolase;
PDBTitle: crystal structure of a putative fumarylacetoacetate2 isomerase/hydrolase from oleispira antarctica
|
| 81 | d1sr9a3 |
|
not modelled |
17.6 |
23 |
Fold:2-isopropylmalate synthase LeuA, allosteric (dimerisation) domain
Superfamily:2-isopropylmalate synthase LeuA, allosteric (dimerisation) domain
Family:2-isopropylmalate synthase LeuA, allosteric (dimerisation) domain |
| 82 | c3peiA_ |
|
not modelled |
16.7 |
18 |
PDB header:hydrolase
Chain: A: PDB Molecule:cytosol aminopeptidase;
PDBTitle: crystal structure of cytosol aminopeptidase from francisella2 tularensis
|
| 83 | d1xmpa_ |
|
not modelled |
15.6 |
22 |
Fold:Flavodoxin-like
Superfamily:N5-CAIR mutase (phosphoribosylaminoimidazole carboxylase, PurE)
Family:N5-CAIR mutase (phosphoribosylaminoimidazole carboxylase, PurE) |
| 84 | c1zmrA_ |
|
not modelled |
15.0 |
17 |
PDB header:transferase
Chain: A: PDB Molecule:phosphoglycerate kinase;
PDBTitle: crystal structure of the e. coli phosphoglycerate kinase
|
| 85 | d1n08a_ |
|
not modelled |
15.0 |
28 |
Fold:Reductase/isomerase/elongation factor common domain
Superfamily:Riboflavin kinase-like
Family:ATP-dependent riboflavin kinase-like |
| 86 | c2gfqC_ |
|
not modelled |
14.9 |
24 |
PDB header:structural genomics, unknown function
Chain: C: PDB Molecule:upf0204 protein ph0006;
PDBTitle: structure of protein of unknown function ph0006 from pyrococcus2 horikoshii
|
| 87 | d2ja9a1 |
|
not modelled |
14.9 |
36 |
Fold:OB-fold
Superfamily:Nucleic acid-binding proteins
Family:Cold shock DNA-binding domain-like |
| 88 | d1vqod1 |
|
not modelled |
14.5 |
26 |
Fold:RL5-like
Superfamily:RL5-like
Family:Ribosomal protein L5 |
| 89 | c2x4lA_ |
|
not modelled |
14.0 |
16 |
PDB header:transport
Chain: A: PDB Molecule:ferric-siderophore receptor protein;
PDBTitle: crystal structure of dese, a ferric-siderophore receptor2 protein from streptomyces coelicolor
|
| 90 | c3rggD_ |
|
not modelled |
12.6 |
15 |
PDB header:lyase
Chain: D: PDB Molecule:phosphoribosylaminoimidazole carboxylase, pure protein;
PDBTitle: crystal structure of treponema denticola pure bound to air
|
| 91 | c2equA_ |
|
not modelled |
12.4 |
33 |
PDB header:protein binding
Chain: A: PDB Molecule:phd finger protein 20-like 1;
PDBTitle: solution structure of the tudor domain of phd finger2 protein 20-like 1
|
| 92 | d1oh1a_ |
|
not modelled |
12.2 |
43 |
Fold:Streptavidin-like
Superfamily:beta-Barrel protease inhibitors
Family:Staphostatin |
| 93 | d1bl0a2 |
|
not modelled |
12.0 |
13 |
Fold:DNA/RNA-binding 3-helical bundle
Superfamily:Homeodomain-like
Family:AraC type transcriptional activator |
| 94 | d2nn6g1 |
|
not modelled |
11.8 |
21 |
Fold:OB-fold
Superfamily:Nucleic acid-binding proteins
Family:Cold shock DNA-binding domain-like |
| 95 | d1hyoa2 |
|
not modelled |
11.8 |
22 |
Fold:FAH
Superfamily:FAH
Family:FAH |
| 96 | c3f6hA_ |
|
not modelled |
11.6 |
18 |
PDB header:transferase
Chain: A: PDB Molecule:alpha-isopropylmalate synthase;
PDBTitle: crystal structure of the regulatory domain of licms in2 complexed with isoleucine - type iii
|
| 97 | d1b12a_ |
|
not modelled |
11.5 |
29 |
Fold:LexA/Signal peptidase
Superfamily:LexA/Signal peptidase
Family:Type 1 signal peptidase |
| 98 | c3trhI_ |
|
not modelled |
11.5 |
20 |
PDB header:lyase
Chain: I: PDB Molecule:phosphoribosylaminoimidazole carboxylase
PDBTitle: structure of a phosphoribosylaminoimidazole carboxylase catalytic2 subunit (pure) from coxiella burnetii
|
| 99 | c1dfwA_ |
|
not modelled |
11.3 |
57 |
PDB header:immune system
Chain: A: PDB Molecule:lung surfactant protein b;
PDBTitle: conformational mapping of the n-terminal segment of2 surfactant protein b in lipid using 13c-enhanced fourier3 transform infrared spectroscopy (ftir)
|