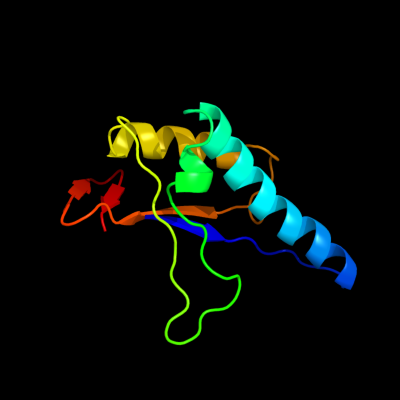
1 c3mcnA_
84.7
18
PDB header: transferaseChain: A: PDB Molecule: 2-amino-4-hydroxy-6-hydroxymethyldihydropteridinePDBTitle: crystal structure of the 6-hyroxymethyl-7,8-dihydropterin2 pyrophosphokinase dihydropteroate synthase bifunctional enzyme from3 francisella tularensis
2 d1f6ya_
67.3
15
Fold: TIM beta/alpha-barrelSuperfamily: Dihydropteroate synthetase-likeFamily: Methyltetrahydrofolate-utiluzing methyltransferases3 c2cg8B_
58.3
38
PDB header: lyase/transferaseChain: B: PDB Molecule: dihydroneopterin aldolase 6-hydroxymethyl-7,8-PDBTitle: the bifunctional dihydroneopterin aldolase 6-hydroxymethyl-2 7,8-dihydropterin synthase from streptococcus pneumoniae
4 d3bofa1
42.5
15
Fold: TIM beta/alpha-barrelSuperfamily: Dihydropteroate synthetase-likeFamily: Methyltetrahydrofolate-utiluzing methyltransferases5 c3a1yG_
40.5
27
PDB header: ribosomal proteinChain: G: PDB Molecule: acidic ribosomal protein p0;PDBTitle: the structure of protein complex
6 d1f9ya_
39.5
20
Fold: Ferredoxin-likeSuperfamily: 6-hydroxymethyl-7,8-dihydropterin pyrophosphokinase, HPPKFamily: 6-hydroxymethyl-7,8-dihydropterin pyrophosphokinase, HPPK7 c3k13A_
37.7
12
PDB header: transferaseChain: A: PDB Molecule: 5-methyltetrahydrofolate-homocysteine methyltransferase;PDBTitle: structure of the pterin-binding domain metr of 5-2 methyltetrahydrofolate-homocysteine methyltransferase from3 bacteroides thetaiotaomicron
8 c2bpbB_
36.7
8
PDB header: oxidoreductaseChain: B: PDB Molecule: sulfite\:cytochrome c oxidoreductase subunit b;PDBTitle: sulfite dehydrogenase from starkeya novella
9 c2qx0A_
36.4
17
PDB header: transferaseChain: A: PDB Molecule: 7,8-dihydro-6-hydroxymethylpterin-PDBTitle: crystal structure of yersinia pestis hppk (ternary complex)
10 c2yciX_
35.9
14
PDB header: transferaseChain: X: PDB Molecule: 5-methyltetrahydrofolate corrinoid/iron sulfur proteinPDBTitle: methyltransferase native
11 d1glua_
35.0
50
Fold: Glucocorticoid receptor-like (DNA-binding domain)Superfamily: Glucocorticoid receptor-like (DNA-binding domain)Family: Nuclear receptor12 c1xyr6_
31.5
33
PDB header: virusChain: 6: PDB Molecule: genome polyprotein, coat protein vp3;PDB Fragment: residues 620-630
PDBTitle: poliovirus 135s cell entry intermediate
13 d1s6la1
30.7
29
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: MerB N-terminal domain-like14 d1dszb_
29.6
15
Fold: Glucocorticoid receptor-like (DNA-binding domain)Superfamily: Glucocorticoid receptor-like (DNA-binding domain)Family: Nuclear receptor15 d1nm8a1
26.8
19
Fold: CoA-dependent acyltransferasesSuperfamily: CoA-dependent acyltransferasesFamily: Choline/Carnitine O-acyltransferase16 d1ndba1
26.5
19
Fold: CoA-dependent acyltransferasesSuperfamily: CoA-dependent acyltransferasesFamily: Choline/Carnitine O-acyltransferase17 d1lo1a_
25.9
46
Fold: Glucocorticoid receptor-like (DNA-binding domain)Superfamily: Glucocorticoid receptor-like (DNA-binding domain)Family: Nuclear receptor18 d1kb6b_
25.5
45
Fold: Glucocorticoid receptor-like (DNA-binding domain)Superfamily: Glucocorticoid receptor-like (DNA-binding domain)Family: Nuclear receptor19 c1r4iA_
24.6
42
PDB header: transcription/dnaChain: A: PDB Molecule: androgen receptor;PDBTitle: crystal structure of androgen receptor dna-binding domain2 bound to a direct repeat response element
20 d1r4ia_
24.6
42
Fold: Glucocorticoid receptor-like (DNA-binding domain)Superfamily: Glucocorticoid receptor-like (DNA-binding domain)Family: Nuclear receptor21 d1cbka_
not modelled
22.7
18
Fold: Ferredoxin-likeSuperfamily: 6-hydroxymethyl-7,8-dihydropterin pyrophosphokinase, HPPKFamily: 6-hydroxymethyl-7,8-dihydropterin pyrophosphokinase, HPPK22 c1no3A_
not modelled
22.1
37
PDB header: oxidoreductaseChain: A: PDB Molecule: lipoxygenase-3;PDBTitle: refined structure of soybean lipoxygenase-3 with 4-nitrocatechol at2 2.15 angstrom resolution
23 d3bnea1
not modelled
22.0
33
Fold: LipoxigenaseSuperfamily: LipoxigenaseFamily: Plant lipoxigenases24 d1hcqa_
not modelled
21.9
33
Fold: Glucocorticoid receptor-like (DNA-binding domain)Superfamily: Glucocorticoid receptor-like (DNA-binding domain)Family: Nuclear receptor25 c1u22A_
not modelled
20.1
17
PDB header: transferaseChain: A: PDB Molecule: 5-methyltetrahydropteroyltriglutamate--PDBTitle: a. thaliana cobalamine independent methionine synthase
26 d1hraa_
not modelled
18.6
17
Fold: Glucocorticoid receptor-like (DNA-binding domain)Superfamily: Glucocorticoid receptor-like (DNA-binding domain)Family: Nuclear receptor27 c2envA_
not modelled
18.6
25
PDB header: transcriptionChain: A: PDB Molecule: peroxisome proliferator-activated receptor delta;PDBTitle: solution sturcture of the c4-type zinc finger domain from2 human peroxisome proliferator-activated receptor delta
28 d1phza1
not modelled
18.5
14
Fold: Ferredoxin-likeSuperfamily: ACT-likeFamily: Phenylalanine metabolism regulatory domain29 c1t7qA_
not modelled
17.9
19
PDB header: transferaseChain: A: PDB Molecule: carnitine acetyltransferase;PDBTitle: crystal structure of the f565a mutant of murine carnitine2 acetyltransferase in complex with carnitine and coa
30 d1ynwa1
not modelled
17.7
42
Fold: Glucocorticoid receptor-like (DNA-binding domain)Superfamily: Glucocorticoid receptor-like (DNA-binding domain)Family: Nuclear receptor31 d1r4ra_
not modelled
17.4
50
Fold: Glucocorticoid receptor-like (DNA-binding domain)Superfamily: Glucocorticoid receptor-like (DNA-binding domain)Family: Nuclear receptor32 c2h4tB_
not modelled
17.4
29
PDB header: transferaseChain: B: PDB Molecule: carnitine o-palmitoyltransferase ii,PDBTitle: crystal structure of rat carnitine palmitoyltransferase ii
33 d1u1ha2
not modelled
16.1
17
Fold: TIM beta/alpha-barrelSuperfamily: UROD/MetE-likeFamily: Cobalamin-independent methionine synthase34 c1nj2A_
not modelled
16.0
23
PDB header: ligaseChain: A: PDB Molecule: proline-trna synthetase;PDBTitle: crystal structure of prolyl-trna synthetase from methanothermobacter2 thermautotrophicus
35 c2e9hA_
not modelled
15.9
12
PDB header: translationChain: A: PDB Molecule: eukaryotic translation initiation factor 5;PDBTitle: solution structure of the eif-5_eif-2b domain from human2 eukaryotic translation initiation factor 5
36 c1qfqB_
not modelled
15.4
31
PDB header: transcription/rnaChain: B: PDB Molecule: 36-mer n-terminal peptide of the n protein;PDBTitle: bacteriophage lambda n-protein-nutboxb-rna complex
37 d1nj1a2
not modelled
15.2
30
Fold: IF3-likeSuperfamily: C-terminal domain of ProRSFamily: C-terminal domain of ProRS38 d1hc7a3
not modelled
15.2
30
Fold: IF3-likeSuperfamily: C-terminal domain of ProRSFamily: C-terminal domain of ProRS39 c2y9zA_
not modelled
15.1
7
PDB header: transcriptionChain: A: PDB Molecule: imitation switch protein 1 (del_atpase);PDBTitle: chromatin remodeling factor isw1a(del_atpase) in dna complex
40 c2eblA_
not modelled
14.5
14
PDB header: transcriptionChain: A: PDB Molecule: coup transcription factor 1;PDBTitle: solution structure of the zinc finger, c4-type domain of2 human coup transcription factor 1
41 d1lata_
not modelled
14.4
50
Fold: Glucocorticoid receptor-like (DNA-binding domain)Superfamily: Glucocorticoid receptor-like (DNA-binding domain)Family: Nuclear receptor42 d1naqa_
not modelled
14.4
11
Fold: Ferredoxin-likeSuperfamily: GlnB-likeFamily: Divalent ion tolerance proteins CutA (CutA1)43 c2fyoA_
not modelled
14.1
29
PDB header: transferaseChain: A: PDB Molecule: carnitine o-palmitoyltransferase ii,PDBTitle: crystal structure of rat carnitine palmitoyltransferase 22 in space group p43212
44 d1rrha1
not modelled
13.7
30
Fold: LipoxigenaseSuperfamily: LipoxigenaseFamily: Plant lipoxigenases45 d1nzaa_
not modelled
13.0
11
Fold: Ferredoxin-likeSuperfamily: GlnB-likeFamily: Divalent ion tolerance proteins CutA (CutA1)46 d1g2ra_
not modelled
12.9
18
Fold: YlxR-likeSuperfamily: YlxR-likeFamily: YlxR-like47 d1z5ye1
not modelled
12.8
19
Fold: Thioredoxin foldSuperfamily: Thioredoxin-likeFamily: Glutathione peroxidase-like48 c1xk8A_
not modelled
12.7
13
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: divalent cation tolerant protein cuta;PDBTitle: divalent cation tolerant protein cuta from homo sapiens2 o60888
49 d2zfha1
not modelled
12.7
13
Fold: Ferredoxin-likeSuperfamily: GlnB-likeFamily: Divalent ion tolerance proteins CutA (CutA1)50 d1vjea_
not modelled
12.4
16
Fold: LuxS/MPP-like metallohydrolaseSuperfamily: LuxS/MPP-like metallohydrolaseFamily: Autoinducer-2 production protein LuxS51 c2x4jA_
not modelled
11.6
67
PDB header: viral proteinChain: A: PDB Molecule: hypothetical protein orf137;PDBTitle: crystal structure of orf137 from pyrobaculum spherical2 virus
52 d1j6xa_
not modelled
11.5
19
Fold: LuxS/MPP-like metallohydrolaseSuperfamily: LuxS/MPP-like metallohydrolaseFamily: Autoinducer-2 production protein LuxS53 c2zfhA_
not modelled
11.5
13
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: cuta;PDBTitle: crystal structure of putative cuta1 from homo sapiens at 2.05a2 resolution
54 d1kr4a_
not modelled
11.5
16
Fold: Ferredoxin-likeSuperfamily: GlnB-likeFamily: Divalent ion tolerance proteins CutA (CutA1)55 c2nuhA_
not modelled
11.5
21
PDB header: unknown functionChain: A: PDB Molecule: periplasmic divalent cation tolerance protein;PDBTitle: crystal structure of cuta from the phytopathgen bacterium xylella2 fastidiosa
56 c2p0gB_
not modelled
11.4
9
PDB header: structural genomics, unknown functionChain: B: PDB Molecule: selenoprotein w-related protein;PDBTitle: crystal structure of selenoprotein w-related protein from2 vibrio cholerae. northeast structural genomics target vcr75
57 d1ukua_
not modelled
11.2
8
Fold: Ferredoxin-likeSuperfamily: GlnB-likeFamily: Divalent ion tolerance proteins CutA (CutA1)58 c2dcuB_
not modelled
11.1
12
PDB header: translationChain: B: PDB Molecule: translation initiation factor 2 beta subunit;PDBTitle: crystal structure of translation initiation factor aif2betagamma2 heterodimer with gdp
59 c1neeA_
not modelled
11.0
16
PDB header: translationChain: A: PDB Molecule: probable translation initiation factor 2 betaPDBTitle: structure of archaeal translation factor aif2beta from2 methanobacterium thermoautrophicum
60 c3dexA_
not modelled
10.9
17
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: sav_2001;PDBTitle: crystal structure of sav_2001 protein from streptomyces2 avermitilis, northeast structural genomics consortium3 target svr107.
61 c2kpaA_
not modelled
10.8
27
PDB header: hydrolaseChain: A: PDB Molecule: arno(375-400);PDBTitle: specific motifs of the v-atpase a2-subunit isoform interact2 with catalytic and regulatory domains of arno
62 c2ojlB_
not modelled
10.6
22
PDB header: structural genomics, unknown functionChain: B: PDB Molecule: hypothetical protein;PDBTitle: crystal structure of q7waf1_borpa from bordetella parapertussis.2 northeast structural genomics target bpr68.
63 c3ialB_
not modelled
10.5
33
PDB header: ligaseChain: B: PDB Molecule: prolyl-trna synthetase;PDBTitle: giardia lamblia prolyl-trna synthetase in complex with prolyl-2 adenylate
64 d2hfva1
not modelled
10.2
28
Fold: Ferredoxin-likeSuperfamily: GlnB-likeFamily: RPA1041-like65 d1j98a_
not modelled
10.1
21
Fold: LuxS/MPP-like metallohydrolaseSuperfamily: LuxS/MPP-like metallohydrolaseFamily: Autoinducer-2 production protein LuxS66 c3bboY_
not modelled
10.1
21
PDB header: ribosomeChain: Y: PDB Molecule: ribosomal protein l28;PDBTitle: homology model for the spinach chloroplast 50s subunit2 fitted to 9.4a cryo-em map of the 70s chlororibosome
67 d1tx2a_
not modelled
9.4
14
Fold: TIM beta/alpha-barrelSuperfamily: Dihydropteroate synthetase-likeFamily: Dihydropteroate synthetase68 c1tx2A_
not modelled
9.4
14
PDB header: transferaseChain: A: PDB Molecule: dhps, dihydropteroate synthase;PDBTitle: dihydropteroate synthetase, with bound inhibitor manic, from bacillus2 anthracis
69 c3mwbA_
not modelled
9.4
19
PDB header: lyaseChain: A: PDB Molecule: prephenate dehydratase;PDBTitle: the crystal structure of prephenate dehydratase in complex with l-phe2 from arthrobacter aurescens to 2.0a
70 d2jz6a1
not modelled
9.2
20
Fold: L28p-likeSuperfamily: L28p-likeFamily: Ribosomal protein L2871 c3ahpA_
not modelled
9.1
14
PDB header: electron transportChain: A: PDB Molecule: cuta1;PDBTitle: crystal structure of stable protein, cuta1, from a psychrotrophic2 bacterium shewanella sp. sib1
72 d1j6wa_
not modelled
9.1
19
Fold: LuxS/MPP-like metallohydrolaseSuperfamily: LuxS/MPP-like metallohydrolaseFamily: Autoinducer-2 production protein LuxS73 d1pbya1
not modelled
9.0
5
Fold: Cytochrome cSuperfamily: Cytochrome cFamily: Quinohemoprotein amine dehydrogenase A chain, domains 1 and 274 c2hfvA_
not modelled
8.7
28
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: hypothetical protein rpa1041;PDBTitle: solution nmr structure of protein rpa1041 from pseudomonas2 aeruginosa. northeast structural genomics consortium3 target pat90.
75 d2es7a1
not modelled
8.4
19
Fold: Thioredoxin foldSuperfamily: Thioredoxin-likeFamily: HyaE-like76 d2nllb_
not modelled
8.3
30
Fold: Glucocorticoid receptor-like (DNA-binding domain)Superfamily: Glucocorticoid receptor-like (DNA-binding domain)Family: Nuclear receptor77 d2qamz1
not modelled
8.3
16
Fold: L28p-likeSuperfamily: L28p-likeFamily: Ribosomal protein L2878 d1dsza_
not modelled
8.2
22
Fold: Glucocorticoid receptor-like (DNA-binding domain)Superfamily: Glucocorticoid receptor-like (DNA-binding domain)Family: Nuclear receptor79 d1k5oa_
not modelled
8.2
39
Fold: Myosin phosphatase inhibitor 17kDa protein, CPI-17Superfamily: Myosin phosphatase inhibitor 17kDa protein, CPI-17Family: Myosin phosphatase inhibitor 17kDa protein, CPI-1780 d1vhfa_
not modelled
8.1
16
Fold: Ferredoxin-likeSuperfamily: GlnB-likeFamily: Divalent ion tolerance proteins CutA (CutA1)81 d1j2na_
not modelled
7.8
36
Fold: Myosin phosphatase inhibitor 17kDa protein, CPI-17Superfamily: Myosin phosphatase inhibitor 17kDa protein, CPI-17Family: Myosin phosphatase inhibitor 17kDa protein, CPI-1782 d1kb2a_
not modelled
7.7
40
Fold: Glucocorticoid receptor-like (DNA-binding domain)Superfamily: Glucocorticoid receptor-like (DNA-binding domain)Family: Nuclear receptor83 d1cita_
not modelled
7.7
44
Fold: Glucocorticoid receptor-like (DNA-binding domain)Superfamily: Glucocorticoid receptor-like (DNA-binding domain)Family: Nuclear receptor84 d1p1la_
not modelled
7.6
8
Fold: Ferredoxin-likeSuperfamily: GlnB-likeFamily: Divalent ion tolerance proteins CutA (CutA1)85 c1a92B_
not modelled
7.5
35
PDB header: leucine zipperChain: B: PDB Molecule: delta antigen;PDBTitle: oligomerization domain of hepatitis delta antigen
86 c3jyw8_
not modelled
7.5
17
PDB header: ribosomeChain: 8: PDB Molecule: 60s ribosomal protein lp0;PDBTitle: structure of the 60s proteins for eukaryotic ribosome based on cryo-em2 map of thermomyces lanuginosus ribosome at 8.9a resolution
87 d1xl7a1
not modelled
7.2
35
Fold: CoA-dependent acyltransferasesSuperfamily: CoA-dependent acyltransferasesFamily: Choline/Carnitine O-acyltransferase88 c2xm5A_
not modelled
6.9
12
PDB header: transferaseChain: A: PDB Molecule: cloq;PDBTitle: structural and mechanistic analysis of the magnesium-2 independent aromatic prenyltransferase cloq from the3 clorobiocin biosynthetic pathway
89 c2zomC_
not modelled
6.9
16
PDB header: unknown functionChain: C: PDB Molecule: protein cuta, chloroplast, putative, expressed;PDBTitle: crystal structure of cuta1 from oryza sativa
90 d2qmwa2
not modelled
6.8
14
Fold: Ferredoxin-likeSuperfamily: ACT-likeFamily: Phenylalanine metabolism regulatory domain91 c2kpbA_
not modelled
6.8
27
PDB header: hydrolaseChain: A: PDB Molecule: arno-p(375-400);PDBTitle: specific motifs of the v-atpase a2-subunit isoform interact2 with catalytic and regulatory domains of arno
92 d1jmxa1
not modelled
6.8
8
Fold: Cytochrome cSuperfamily: Cytochrome cFamily: Quinohemoprotein amine dehydrogenase A chain, domains 1 and 293 d1ef4a_
not modelled
6.7
50
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: RNA polymerase subunit RPB10Family: RNA polymerase subunit RPB1094 c3swfA_
not modelled
6.7
17
PDB header: transport proteinChain: A: PDB Molecule: cgmp-gated cation channel alpha-1;PDBTitle: cnga1 621-690 containing clz domain
95 c1yx7A_
not modelled
6.5
12
PDB header: metal binding proteinChain: A: PDB Molecule: calsensin;PDBTitle: nmr structure of calsensin, energy minimized average2 structure.
96 d2d8va1
not modelled
6.4
38
Fold: B-box zinc-binding domainSuperfamily: B-box zinc-binding domainFamily: B-box zinc-binding domain97 d1yuza2
not modelled
6.2
14
Fold: Rubredoxin-likeSuperfamily: Rubredoxin-likeFamily: Rubredoxin98 c1lj2B_
not modelled
6.1
33
PDB header: viral protein/ translationChain: B: PDB Molecule: nonstructural rna-binding protein 34;PDBTitle: recognition of eif4g by rotavirus nsp3 reveals a basis for2 mrna circularization
99 d1t1ua1
not modelled
6.1
25
Fold: CoA-dependent acyltransferasesSuperfamily: CoA-dependent acyltransferasesFamily: Choline/Carnitine O-acyltransferase