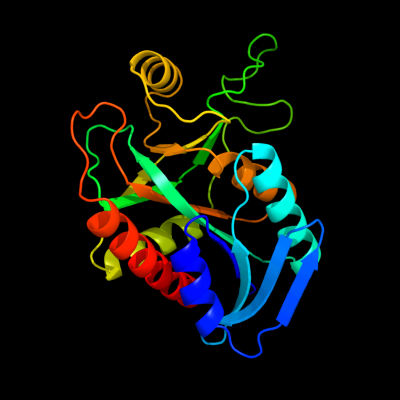
| 1 | c3bl6A_
|
|
 |
100.0 |
55 |
PDB header:hydrolase
Chain: A: PDB Molecule:5'-methylthioadenosine nucleosidase/s-
PDBTitle: crystal structure of staphylococcus aureus 5'-2 methylthioadenosine/s-adenosylhomocysteine nucleosidase in3 complex with formycin a
|
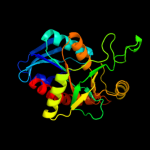
| 2 | c1zosE_
|
|
 |
100.0 |
41 |
PDB header:hydrolase
Chain: E: PDB Molecule:5'-methylthioadenosine / s-adenosylhomocysteine
PDBTitle: structure of 5'-methylthionadenosine/s-adenosylhomocysteine2 nucleosidase from s. pneumoniae with a transition-state3 inhibitor mt-imma
|
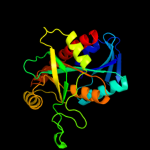
| 3 | d1jysa_
|
|
 |
100.0 |
100 |
Fold:Phosphorylase/hydrolase-like
Superfamily:Purine and uridine phosphorylases
Family:Purine and uridine phosphorylases |
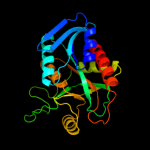
| 4 | c3eeiA_
|
|
 |
100.0 |
49 |
PDB header:hydrolase
Chain: A: PDB Molecule:5-methylthioadenosine nucleosidase/s-
PDBTitle: crystal structure of 5'-methylthioadenosine/s-2 adenosylhomocysteine nucleosidase from neisseria3 meningitidis in complex with methylthio-immucillin-a
|
| 5 | c3dp9A_
|
|
 |
100.0 |
61 |
PDB header:hydrolase
Chain: A: PDB Molecule:mta/sah nucleosidase;
PDBTitle: crystal structure of vibrio cholerae 5'-methylthioadenosine/s-adenosyl2 homocysteine nucleosidase (mtan) complexed with butylthio-dadme-3 immucillin a
|
| 6 | c3nm5B_
|
|
 |
100.0 |
35 |
PDB header:hydrolase
Chain: B: PDB Molecule:mta/sah nucleosidase;
PDBTitle: helicobacter pylori mtan complexed with formycin a
|
| 7 | c2h8gA_
|
|
 |
100.0 |
24 |
PDB header:hydrolase
Chain: A: PDB Molecule:5'-methylthioadenosine nucleosidase;
PDBTitle: 5'-methylthioadenosine nucleosidase from arabidopsis2 thaliana
|
| 8 | c3bsfB_
|
|
 |
100.0 |
23 |
PDB header:hydrolase
Chain: B: PDB Molecule:at4g34840;
PDBTitle: crystal structure of the mta/sah nucleosidase
|
| 9 | c1z34A_
|
|
 |
100.0 |
21 |
PDB header:transferase
Chain: A: PDB Molecule:purine nucleoside phosphorylase;
PDBTitle: crystal structure of trichomonas vaginalis purine nucleoside2 phosphorylase complexed with 2-fluoro-2'-deoxyadenosine
|
| 10 | d1rxya_
|
|
 |
100.0 |
16 |
Fold:Phosphorylase/hydrolase-like
Superfamily:Purine and uridine phosphorylases
Family:Purine and uridine phosphorylases |
| 11 | d1je0a_
|
|
 |
100.0 |
21 |
Fold:Phosphorylase/hydrolase-like
Superfamily:Purine and uridine phosphorylases
Family:Purine and uridine phosphorylases |
| 12 | d1vhwa_
|
|
 |
100.0 |
18 |
Fold:Phosphorylase/hydrolase-like
Superfamily:Purine and uridine phosphorylases
Family:Purine and uridine phosphorylases |
| 13 | c1nw4C_
|
|
 |
100.0 |
19 |
PDB header:transferase
Chain: C: PDB Molecule:uridine phosphorylase, putative;
PDBTitle: crystal structure of plasmodium falciparum purine nucleoside2 phosphorylase in complex with immh and sulfate
|
| 14 | d1odka_
|
|
 |
100.0 |
22 |
Fold:Phosphorylase/hydrolase-like
Superfamily:Purine and uridine phosphorylases
Family:Purine and uridine phosphorylases |
| 15 | d1q1ga_
|
|
 |
100.0 |
19 |
Fold:Phosphorylase/hydrolase-like
Superfamily:Purine and uridine phosphorylases
Family:Purine and uridine phosphorylases |
| 16 | d2ac7a1
|
|
 |
100.0 |
19 |
Fold:Phosphorylase/hydrolase-like
Superfamily:Purine and uridine phosphorylases
Family:Purine and uridine phosphorylases |
| 17 | d1k9sa_
|
|
 |
100.0 |
18 |
Fold:Phosphorylase/hydrolase-like
Superfamily:Purine and uridine phosphorylases
Family:Purine and uridine phosphorylases |
| 18 | c3mb8A_
|
|
 |
100.0 |
18 |
PDB header:transferase
Chain: A: PDB Molecule:purine nucleoside phosphorylase;
PDBTitle: crystal structure of purine nucleoside phosphorylase from toxoplasma2 gondii in complex with immucillin-h
|
| 19 | c3tl6B_
|
|
 |
100.0 |
27 |
PDB header:transferase
Chain: B: PDB Molecule:purine nucleoside phosphorylase;
PDBTitle: crystal structure of purine nucleoside phosphorylase from entamoeba2 histolytica
|
| 20 | c3qpbB_
|
|
 |
100.0 |
19 |
PDB header:transferase
Chain: B: PDB Molecule:uridine phosphorylase;
PDBTitle: crystal structure of streptococcus pyogenes uridine phosphorylase2 reveals a subclass of the np-i superfamily
|
| 21 | d1ybfa_ |
|
not modelled |
100.0 |
16 |
Fold:Phosphorylase/hydrolase-like
Superfamily:Purine and uridine phosphorylases
Family:Purine and uridine phosphorylases |
| 22 | d1t8sa_ |
|
not modelled |
100.0 |
19 |
Fold:Phosphorylase/hydrolase-like
Superfamily:Purine and uridine phosphorylases
Family:Purine and uridine phosphorylases |
| 23 | d1v4na_ |
|
not modelled |
100.0 |
17 |
Fold:Phosphorylase/hydrolase-like
Superfamily:Purine and uridine phosphorylases
Family:Purine and uridine phosphorylases |
| 24 | c3khsB_ |
|
not modelled |
100.0 |
19 |
PDB header:hydrolase
Chain: B: PDB Molecule:purine nucleoside phosphorylase;
PDBTitle: crystal structure of grouper iridovirus purine nucleoside2 phosphorylase
|
| 25 | c2xrfA_ |
|
not modelled |
100.0 |
16 |
PDB header:transferase
Chain: A: PDB Molecule:uridine phosphorylase 2;
PDBTitle: crystal structure of human uridine phosphorylase 2
|
| 26 | c1wtaA_ |
|
not modelled |
100.0 |
16 |
PDB header:transferase
Chain: A: PDB Molecule:5'-methylthioadenosine phosphorylase;
PDBTitle: crystal structure of 5'-deoxy-5'-methylthioadenosine from aeropyrum2 pernix (r32 form)
|
| 27 | c3ozbF_ |
|
not modelled |
100.0 |
14 |
PDB header:transferase
Chain: F: PDB Molecule:methylthioadenosine phosphorylase;
PDBTitle: crystal structure of 5'-methylthioinosine phosphorylase from2 psedomonas aeruginosa in complex with hypoxanthine
|
| 28 | c1tcvB_ |
|
not modelled |
100.0 |
18 |
PDB header:transferase
Chain: B: PDB Molecule:purine-nucleoside phosphorylase;
PDBTitle: crystal structure of the purine nucleoside phosphorylase2 from schistosoma mansoni in complex with non-detergent3 sulfobetaine 195 and acetate
|
| 29 | d1cb0a_ |
|
not modelled |
100.0 |
16 |
Fold:Phosphorylase/hydrolase-like
Superfamily:Purine and uridine phosphorylases
Family:Purine and uridine phosphorylases |
| 30 | c3eufC_ |
|
not modelled |
100.0 |
18 |
PDB header:transferase
Chain: C: PDB Molecule:uridine phosphorylase 1;
PDBTitle: crystal structure of bau-bound human uridine phosphorylase 1
|
| 31 | d1vmka_ |
|
not modelled |
100.0 |
14 |
Fold:Phosphorylase/hydrolase-like
Superfamily:Purine and uridine phosphorylases
Family:Purine and uridine phosphorylases |
| 32 | c3bjeA_ |
|
not modelled |
100.0 |
16 |
PDB header:transferase
Chain: A: PDB Molecule:nucleoside phosphorylase, putative;
PDBTitle: crystal structure of trypanosoma brucei nucleoside phosphorylase shows2 uridine phosphorylase activity
|
| 33 | c2p4sA_ |
|
not modelled |
100.0 |
16 |
PDB header:transferase
Chain: A: PDB Molecule:purine nucleoside phosphorylase;
PDBTitle: structure of purine nucleoside phosphorylase from anopheles gambiae in2 complex with dadme-immh
|
| 34 | c3la8A_ |
|
not modelled |
100.0 |
15 |
PDB header:transferase
Chain: A: PDB Molecule:putative purine nucleoside phosphorylase;
PDBTitle: the crystal structure of smu.1229 from streptococcus mutans ua159
|
| 35 | c1yr3A_ |
|
not modelled |
100.0 |
18 |
PDB header:transferase
Chain: A: PDB Molecule:xanthosine phosphorylase;
PDBTitle: escherichia coli purine nucleoside phosphorylase ii, the2 product of the xapa gene
|
| 36 | d3bgsa1 |
|
not modelled |
100.0 |
17 |
Fold:Phosphorylase/hydrolase-like
Superfamily:Purine and uridine phosphorylases
Family:Purine and uridine phosphorylases |
| 37 | d1g2oa_ |
|
not modelled |
99.9 |
13 |
Fold:Phosphorylase/hydrolase-like
Superfamily:Purine and uridine phosphorylases
Family:Purine and uridine phosphorylases |
| 38 | c3ggsA_ |
|
not modelled |
99.9 |
19 |
PDB header:transferase
Chain: A: PDB Molecule:purine nucleoside phosphorylase;
PDBTitle: human purine nucleoside phosphorylase double mutant e201q,n243d2 complexed with 2-fluoro-2'-deoxyadenosine
|
| 39 | d3pnpa_ |
|
not modelled |
99.9 |
16 |
Fold:Phosphorylase/hydrolase-like
Superfamily:Purine and uridine phosphorylases
Family:Purine and uridine phosphorylases |
| 40 | d1qe5a_ |
|
not modelled |
99.9 |
16 |
Fold:Phosphorylase/hydrolase-like
Superfamily:Purine and uridine phosphorylases
Family:Purine and uridine phosphorylases |
| 41 | d1a2za_ |
|
not modelled |
60.9 |
24 |
Fold:Phosphorylase/hydrolase-like
Superfamily:Pyrrolidone carboxyl peptidase (pyroglutamate aminopeptidase)
Family:Pyrrolidone carboxyl peptidase (pyroglutamate aminopeptidase) |
| 42 | c3giuA_ |
|
not modelled |
50.7 |
19 |
PDB header:hydrolase
Chain: A: PDB Molecule:pyrrolidone-carboxylate peptidase;
PDBTitle: 1.25 angstrom crystal structure of pyrrolidone-carboxylate peptidase2 (pcp) from staphylococcus aureus
|
| 43 | c3lacA_ |
|
not modelled |
39.8 |
17 |
PDB header:hydrolase
Chain: A: PDB Molecule:pyrrolidone-carboxylate peptidase;
PDBTitle: crystal structure of bacillus anthracis pyrrolidone-carboxylate2 peptidase, pcp
|
| 44 | d1iofa_ |
|
not modelled |
35.0 |
24 |
Fold:Phosphorylase/hydrolase-like
Superfamily:Pyrrolidone carboxyl peptidase (pyroglutamate aminopeptidase)
Family:Pyrrolidone carboxyl peptidase (pyroglutamate aminopeptidase) |
| 45 | d1auga_ |
|
not modelled |
22.7 |
17 |
Fold:Phosphorylase/hydrolase-like
Superfamily:Pyrrolidone carboxyl peptidase (pyroglutamate aminopeptidase)
Family:Pyrrolidone carboxyl peptidase (pyroglutamate aminopeptidase) |
| 46 | c2ywjA_ |
|
not modelled |
20.5 |
21 |
PDB header:transferase
Chain: A: PDB Molecule:glutamine amidotransferase subunit pdxt;
PDBTitle: crystal structure of uncharacterized conserved protein from2 methanocaldococcus jannaschii
|
| 47 | d1vb5a_ |
|
not modelled |
16.6 |
33 |
Fold:NagB/RpiA/CoA transferase-like
Superfamily:NagB/RpiA/CoA transferase-like
Family:IF2B-like |
| 48 | d2nv0a1 |
|
not modelled |
15.6 |
18 |
Fold:Flavodoxin-like
Superfamily:Class I glutamine amidotransferase-like
Family:Class I glutamine amidotransferases (GAT) |
| 49 | d1t9ka_ |
|
not modelled |
15.2 |
28 |
Fold:NagB/RpiA/CoA transferase-like
Superfamily:NagB/RpiA/CoA transferase-like
Family:IF2B-like |
| 50 | c2equA_ |
|
not modelled |
14.8 |
30 |
PDB header:protein binding
Chain: A: PDB Molecule:phd finger protein 20-like 1;
PDBTitle: solution structure of the tudor domain of phd finger2 protein 20-like 1
|
| 51 | c2yvkA_ |
|
not modelled |
14.7 |
28 |
PDB header:isomerase
Chain: A: PDB Molecule:methylthioribose-1-phosphate isomerase;
PDBTitle: crystal structure of 5-methylthioribose 1-phosphate2 isomerase product complex from bacillus subtilis
|
| 52 | d2a0ua1 |
|
not modelled |
14.1 |
17 |
Fold:NagB/RpiA/CoA transferase-like
Superfamily:NagB/RpiA/CoA transferase-like
Family:IF2B-like |
| 53 | d1fdra2 |
|
not modelled |
13.9 |
10 |
Fold:Ferredoxin reductase-like, C-terminal NADP-linked domain
Superfamily:Ferredoxin reductase-like, C-terminal NADP-linked domain
Family:Reductases |
| 54 | d1t5oa_ |
|
not modelled |
12.8 |
28 |
Fold:NagB/RpiA/CoA transferase-like
Superfamily:NagB/RpiA/CoA transferase-like
Family:IF2B-like |
| 55 | c1yt5A_ |
|
not modelled |
12.2 |
22 |
PDB header:transferase
Chain: A: PDB Molecule:inorganic polyphosphate/atp-nad kinase;
PDBTitle: crystal structure of nad kinase from thermotoga maritima
|
| 56 | c3cwcB_ |
|
not modelled |
12.1 |
14 |
PDB header:transferase
Chain: B: PDB Molecule:putative glycerate kinase 2;
PDBTitle: crystal structure of putative glycerate kinase 2 from salmonella2 typhimurium lt2
|
| 57 | d1txga2 |
|
not modelled |
11.1 |
50 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:6-phosphogluconate dehydrogenase-like, N-terminal domain |
| 58 | d2f1ka2 |
|
not modelled |
10.2 |
63 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:6-phosphogluconate dehydrogenase-like, N-terminal domain |
| 59 | c1tvmA_ |
|
not modelled |
9.9 |
8 |
PDB header:transferase
Chain: A: PDB Molecule:pts system, galactitol-specific iib component;
PDBTitle: nmr structure of enzyme gatb of the galactitol-specific2 phosphoenolpyruvate-dependent phosphotransferase system
|
| 60 | c3a11D_ |
|
not modelled |
9.4 |
17 |
PDB header:isomerase
Chain: D: PDB Molecule:translation initiation factor eif-2b, delta
PDBTitle: crystal structure of ribose-1,5-bisphosphate isomerase from2 thermococcus kodakaraensis kod1
|
| 61 | d1jfla1 |
|
not modelled |
9.3 |
42 |
Fold:ATC-like
Superfamily:Aspartate/glutamate racemase
Family:Aspartate/glutamate racemase |
| 62 | c1w2wJ_ |
|
not modelled |
9.1 |
26 |
PDB header:isomerase
Chain: J: PDB Molecule:5-methylthioribose-1-phosphate isomerase;
PDBTitle: crystal structure of yeast ypr118w, a methylthioribose-1-2 phosphate isomerase related to regulatory eif2b subunits
|
| 63 | c2pjkA_ |
|
not modelled |
8.7 |
17 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:178aa long hypothetical molybdenum cofactor
PDBTitle: structure of hypothetical molybdenum cofactor biosynthesis2 protein b from sulfolobus tokodaii
|
| 64 | d1eo1a_ |
|
not modelled |
8.7 |
10 |
Fold:Ribonuclease H-like motif
Superfamily:Nitrogenase accessory factor-like
Family:MTH1175-like |
| 65 | d1vola1 |
|
not modelled |
8.4 |
15 |
Fold:Cyclin-like
Superfamily:Cyclin-like
Family:Transcription factor IIB (TFIIB), core domain |
| 66 | c2qneA_ |
|
not modelled |
7.9 |
22 |
PDB header:transferase
Chain: A: PDB Molecule:putative methyltransferase;
PDBTitle: crystal structure of putative methyltransferase (zp_00558420.1) from2 desulfitobacterium hafniense y51 at 2.30 a resolution
|
| 67 | d1cqxa3 |
|
not modelled |
7.7 |
18 |
Fold:Ferredoxin reductase-like, C-terminal NADP-linked domain
Superfamily:Ferredoxin reductase-like, C-terminal NADP-linked domain
Family:Flavohemoglobin, C-terminal domain |
| 68 | d1gkub3 |
|
not modelled |
7.7 |
13 |
Fold:Prokaryotic type I DNA topoisomerase
Superfamily:Prokaryotic type I DNA topoisomerase
Family:Prokaryotic type I DNA topoisomerase |
| 69 | c3peiA_ |
|
not modelled |
7.7 |
27 |
PDB header:hydrolase
Chain: A: PDB Molecule:cytosol aminopeptidase;
PDBTitle: crystal structure of cytosol aminopeptidase from francisella2 tularensis
|
| 70 | d2bm8a1 |
|
not modelled |
7.6 |
22 |
Fold:S-adenosyl-L-methionine-dependent methyltransferases
Superfamily:S-adenosyl-L-methionine-dependent methyltransferases
Family:CmcI-like |
| 71 | c2wfbA_ |
|
not modelled |
7.3 |
10 |
PDB header:biosynthetic protein
Chain: A: PDB Molecule:putative uncharacterized protein orp;
PDBTitle: high resolution structure of the apo form of the orange2 protein (orp) from desulfovibrio gigas
|
| 72 | d1gvha3 |
|
not modelled |
7.2 |
16 |
Fold:Ferredoxin reductase-like, C-terminal NADP-linked domain
Superfamily:Ferredoxin reductase-like, C-terminal NADP-linked domain
Family:Flavohemoglobin, C-terminal domain |
| 73 | d1dxya2 |
|
not modelled |
7.2 |
29 |
Fold:Flavodoxin-like
Superfamily:Formate/glycerate dehydrogenase catalytic domain-like
Family:Formate/glycerate dehydrogenases, substrate-binding domain |
| 74 | d1iu8a_ |
|
not modelled |
7.1 |
25 |
Fold:Phosphorylase/hydrolase-like
Superfamily:Pyrrolidone carboxyl peptidase (pyroglutamate aminopeptidase)
Family:Pyrrolidone carboxyl peptidase (pyroglutamate aminopeptidase) |
| 75 | d1o6ca_ |
|
not modelled |
6.8 |
17 |
Fold:UDP-Glycosyltransferase/glycogen phosphorylase
Superfamily:UDP-Glycosyltransferase/glycogen phosphorylase
Family:UDP-N-acetylglucosamine 2-epimerase |
| 76 | d2fug61 |
|
not modelled |
6.6 |
17 |
Fold:HydA/Nqo6-like
Superfamily:HydA/Nqo6-like
Family:Nq06-like |
| 77 | d1sqsa_ |
|
not modelled |
6.5 |
11 |
Fold:Flavodoxin-like
Superfamily:Flavoproteins
Family:Hypothetical protein SP1951 |
| 78 | d1rdua_ |
|
not modelled |
6.4 |
8 |
Fold:Ribonuclease H-like motif
Superfamily:Nitrogenase accessory factor-like
Family:MTH1175-like |
| 79 | d1l5xa_ |
|
not modelled |
6.3 |
12 |
Fold:SurE-like
Superfamily:SurE-like
Family:SurE-like |
| 80 | d1ks9a2 |
|
not modelled |
6.0 |
50 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:6-phosphogluconate dehydrogenase-like, N-terminal domain |
| 81 | d1a8pa2 |
|
not modelled |
5.9 |
10 |
Fold:Ferredoxin reductase-like, C-terminal NADP-linked domain
Superfamily:Ferredoxin reductase-like, C-terminal NADP-linked domain
Family:Reductases |
| 82 | c2f1kD_ |
|
not modelled |
5.9 |
63 |
PDB header:oxidoreductase
Chain: D: PDB Molecule:prephenate dehydrogenase;
PDBTitle: crystal structure of synechocystis arogenate dehydrogenase
|
| 83 | d1wd5a_ |
|
not modelled |
5.8 |
18 |
Fold:PRTase-like
Superfamily:PRTase-like
Family:Phosphoribosyltransferases (PRTases) |
| 84 | d1mv8a2 |
|
not modelled |
5.8 |
50 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:6-phosphogluconate dehydrogenase-like, N-terminal domain |
| 85 | c1olsB_ |
|
not modelled |
5.8 |
11 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:2-oxoisovalerate dehydrogenase beta subunit;
PDBTitle: roles of his291-alpha and his146-beta' in the reductive2 acylation reaction catalyzed by human branched-chain3 alpha-ketoacid dehydrogenase
|
| 86 | d1j0aa_ |
|
not modelled |
5.7 |
11 |
Fold:Tryptophan synthase beta subunit-like PLP-dependent enzymes
Superfamily:Tryptophan synthase beta subunit-like PLP-dependent enzymes
Family:Tryptophan synthase beta subunit-like PLP-dependent enzymes |
| 87 | d1a5za1 |
|
not modelled |
5.7 |
75 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:LDH N-terminal domain-like |
| 88 | d1gv0a1 |
|
not modelled |
5.6 |
75 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:LDH N-terminal domain-like |
| 89 | d1pvda2 |
|
not modelled |
5.4 |
15 |
Fold:Thiamin diphosphate-binding fold (THDP-binding)
Superfamily:Thiamin diphosphate-binding fold (THDP-binding)
Family:Pyruvate oxidase and decarboxylase Pyr module |
| 90 | d1zpda2 |
|
not modelled |
5.4 |
18 |
Fold:Thiamin diphosphate-binding fold (THDP-binding)
Superfamily:Thiamin diphosphate-binding fold (THDP-binding)
Family:Pyruvate oxidase and decarboxylase Pyr module |
| 91 | d1ojua1 |
|
not modelled |
5.3 |
63 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:LDH N-terminal domain-like |
| 92 | c3snrA_ |
|
not modelled |
5.3 |
22 |
PDB header:transport protein
Chain: A: PDB Molecule:extracellular ligand-binding receptor;
PDBTitle: rpd_1889 protein, an extracellular ligand-binding receptor from2 rhodopseudomonas palustris.
|
| 93 | c3ecsD_ |
|
not modelled |
5.3 |
26 |
PDB header:translation
Chain: D: PDB Molecule:translation initiation factor eif-2b subunit
PDBTitle: crystal structure of human eif2b alpha
|
| 94 | c3fysA_ |
|
not modelled |
5.2 |
29 |
PDB header:fatty acid-binding protein
Chain: A: PDB Molecule:protein degv;
PDBTitle: crystal structure of degv, a fatty acid binding protein2 from bacillus subtilis
|
| 95 | c2gf2B_ |
|
not modelled |
5.1 |
50 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:3-hydroxyisobutyrate dehydrogenase;
PDBTitle: crystal structure of human hydroxyisobutyrate dehydrogenase
|
| 96 | d2bfdb1 |
|
not modelled |
5.1 |
11 |
Fold:Thiamin diphosphate-binding fold (THDP-binding)
Superfamily:Thiamin diphosphate-binding fold (THDP-binding)
Family:Branched-chain alpha-keto acid dehydrogenase Pyr module |