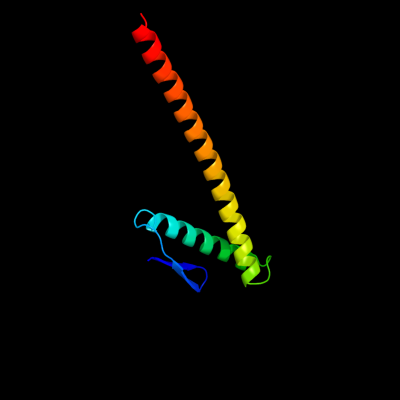
| 1 | d1t3ua_
|
|
 |
99.9 |
27 |
Fold:Cell division protein ZapA-like
Superfamily:Cell division protein ZapA-like
Family:Cell division protein ZapA-like |
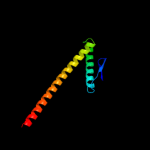
| 2 | c3hnwB_
|
|
 |
99.8 |
12 |
PDB header:structural genomics, unknown function
Chain: B: PDB Molecule:uncharacterized protein;
PDBTitle: crystal structure of a basic coiled-coil protein of unknown function2 from eubacterium eligens atcc 27750
|
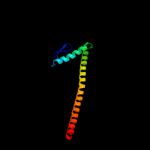
| 3 | c2ns5A_
|
|
 |
64.8 |
11 |
PDB header:signaling protein
Chain: A: PDB Molecule:partitioning-defective 3 homolog;
PDBTitle: the conserved n-terminal domain of par-3 adopts a novel pb1-2 like structure required for par-3 oligomerization and3 apical membrane localization
|
| 4 | c3hrdH_
|
|
 |
64.2 |
13 |
PDB header:oxidoreductase
Chain: H: PDB Molecule:nicotinate dehydrogenase small fes subunit;
PDBTitle: crystal structure of nicotinate dehydrogenase
|
| 5 | d1t3qa2
|
|
 |
52.3 |
16 |
Fold:beta-Grasp (ubiquitin-like)
Superfamily:2Fe-2S ferredoxin-like
Family:2Fe-2S ferredoxin domains from multidomain proteins |
| 6 | d1rm6c2
|
|
 |
50.0 |
18 |
Fold:beta-Grasp (ubiquitin-like)
Superfamily:2Fe-2S ferredoxin-like
Family:2Fe-2S ferredoxin domains from multidomain proteins |
| 7 | d1vlba2
|
|
 |
45.8 |
11 |
Fold:beta-Grasp (ubiquitin-like)
Superfamily:2Fe-2S ferredoxin-like
Family:2Fe-2S ferredoxin domains from multidomain proteins |
| 8 | d1ffva2
|
|
 |
44.1 |
10 |
Fold:beta-Grasp (ubiquitin-like)
Superfamily:2Fe-2S ferredoxin-like
Family:2Fe-2S ferredoxin domains from multidomain proteins |
| 9 | c2x7aB_
|
|
 |
42.3 |
21 |
PDB header:immune system
Chain: B: PDB Molecule:bone marrow stromal antigen 2;
PDBTitle: structural basis of hiv-1 tethering to membranes by the2 bst2-tetherin ectodomain
|
| 10 | d1n62a2
|
|
 |
40.6 |
16 |
Fold:beta-Grasp (ubiquitin-like)
Superfamily:2Fe-2S ferredoxin-like
Family:2Fe-2S ferredoxin domains from multidomain proteins |
| 11 | c1t3qD_
|
|
 |
38.8 |
16 |
PDB header:oxidoreductase
Chain: D: PDB Molecule:quinoline 2-oxidoreductase small subunit;
PDBTitle: crystal structure of quinoline 2-oxidoreductase from pseudomonas2 putida 86
|
| 12 | c1rm6F_
|
|
 |
37.4 |
18 |
PDB header:oxidoreductase
Chain: F: PDB Molecule:4-hydroxybenzoyl-coa reductase gamma subunit;
PDBTitle: structure of 4-hydroxybenzoyl-coa reductase from thauera2 aromatica
|
| 13 | d1dgja2
|
|
 |
37.4 |
15 |
Fold:beta-Grasp (ubiquitin-like)
Superfamily:2Fe-2S ferredoxin-like
Family:2Fe-2S ferredoxin domains from multidomain proteins |
| 14 | c1x4rA_
|
|
 |
33.5 |
19 |
PDB header:apoptosis
Chain: A: PDB Molecule:parp14 protein;
PDBTitle: solution structure of wwe domain in parp14 protein
|
| 15 | c1ffuA_
|
|
 |
32.7 |
10 |
PDB header:hydrolase
Chain: A: PDB Molecule:cuts, iron-sulfur protein of carbon monoxide
PDBTitle: carbon monoxide dehydrogenase from hydrogenophaga2 pseudoflava which lacks the mo-pyranopterin moiety of the3 molybdenum cofactor
|
| 16 | d3c8ya2
|
|
 |
28.0 |
27 |
Fold:beta-Grasp (ubiquitin-like)
Superfamily:2Fe-2S ferredoxin-like
Family:2Fe-2S ferredoxin domains from multidomain proteins |
| 17 | c1n60D_
|
|
 |
27.3 |
16 |
PDB header:oxidoreductase
Chain: D: PDB Molecule:carbon monoxide dehydrogenase small chain;
PDBTitle: crystal structure of the cu,mo-co dehydrogenase (codh); cyanide-2 inactivated form
|
| 18 | d2fug33
|
|
 |
22.5 |
23 |
Fold:beta-Grasp (ubiquitin-like)
Superfamily:2Fe-2S ferredoxin-like
Family:2Fe-2S ferredoxin domains from multidomain proteins |
| 19 | d1hyea2
|
|
 |
22.0 |
13 |
Fold:LDH C-terminal domain-like
Superfamily:LDH C-terminal domain-like
Family:Lactate & malate dehydrogenases, C-terminal domain |
| 20 | d1iuea_
|
|
 |
21.4 |
12 |
Fold:beta-Grasp (ubiquitin-like)
Superfamily:2Fe-2S ferredoxin-like
Family:2Fe-2S ferredoxin-related |
| 21 | c3htkA_ |
|
not modelled |
20.2 |
22 |
PDB header:recombination/replication/ligase
Chain: A: PDB Molecule:structural maintenance of chromosomes protein 5;
PDBTitle: crystal structure of mms21 and smc5 complex
|
| 22 | d2oq0a1 |
|
not modelled |
19.9 |
16 |
Fold:OB-fold
Superfamily:HIN-2000 domain-like
Family:HIN-200/IF120x domain |
| 23 | c2fugC_ |
|
not modelled |
17.4 |
24 |
PDB header:oxidoreductase
Chain: C: PDB Molecule:nadh-quinone oxidoreductase chain 3;
PDBTitle: crystal structure of the hydrophilic domain of respiratory complex i2 from thermus thermophilus
|
| 24 | d1exbe_ |
|
not modelled |
16.7 |
29 |
Fold:POZ domain
Superfamily:POZ domain
Family:Tetramerization domain of potassium channels |
| 25 | c3dryA_ |
|
not modelled |
16.4 |
13 |
PDB header:unknown function
Chain: A: PDB Molecule:btb/poz domain-containing protein kctd5;
PDBTitle: x-ray crystal structure of human kctd5 protein crystallized in low-2 salt buffer
|
| 26 | d1qdva_ |
|
not modelled |
16.3 |
29 |
Fold:POZ domain
Superfamily:POZ domain
Family:Tetramerization domain of potassium channels |
| 27 | d1t1da_ |
|
not modelled |
16.0 |
21 |
Fold:POZ domain
Superfamily:POZ domain
Family:Tetramerization domain of potassium channels |
| 28 | d1t95a2 |
|
not modelled |
15.8 |
18 |
Fold:FYSH domain
Superfamily:FYSH domain
Family:Hypothetical protein AF0491, N-terminal domain |
| 29 | d2pw9a1 |
|
not modelled |
15.8 |
17 |
Fold:Cytidine deaminase-like
Superfamily:Cytidine deaminase-like
Family:FdhD/NarQ |
| 30 | c2oq0D_ |
|
not modelled |
15.7 |
16 |
PDB header:protein binding
Chain: D: PDB Molecule:gamma-interferon-inducible protein ifi-16;
PDBTitle: crystal structure of the first hin-200 domain of interferon-inducible2 protein 16
|
| 31 | d1dsxa_ |
|
not modelled |
15.6 |
33 |
Fold:POZ domain
Superfamily:POZ domain
Family:Tetramerization domain of potassium channels |
| 32 | c2kanA_ |
|
not modelled |
15.3 |
11 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:uncharacterized protein ar3433a;
PDBTitle: solution nmr structure of ubiquitin-like domain of2 arabidopsis thaliana protein at2g32350. northeast3 structural genomics consortium target ar3433a
|
| 33 | c2it3B_ |
|
not modelled |
14.7 |
19 |
PDB header:structural genomics, unknown function
Chain: B: PDB Molecule:upf0130 protein ph1069;
PDBTitle: structure of ph1069 protein from pyrococcus horikoshii
|
| 34 | c3b6yB_ |
|
not modelled |
14.0 |
21 |
PDB header:protein binding
Chain: B: PDB Molecule:gamma-interferon-inducible protein ifi-16;
PDBTitle: crystal structure of the second hin-200 domain of interferon-inducible2 protein 16
|
| 35 | c1c4cA_ |
|
not modelled |
13.8 |
27 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:protein (fe-only hydrogenase);
PDBTitle: binding of exogenously added carbon monoxide at the active2 site of the fe-only hydrogenase (cpi) from clostridium3 pasteurianum
|
| 36 | c2kd0A_ |
|
not modelled |
13.7 |
23 |
PDB header:signaling protein
Chain: A: PDB Molecule:lrr repeats and ubiquitin-like domain-containing
PDBTitle: nmr solution structure of o64736 protein from arabidopsis2 thaliana. northeast structural genomics consortium mega3 target ar3445a
|
| 37 | c2x48B_ |
|
not modelled |
12.9 |
22 |
PDB header:viral protein
Chain: B: PDB Molecule:cag38821;
PDBTitle: orf 55 from sulfolobus islandicus rudivirus 1
|
| 38 | c3cwiA_ |
|
not modelled |
12.6 |
13 |
PDB header:biosynthetic protein
Chain: A: PDB Molecule:thiamine-biosynthesis protein this;
PDBTitle: crystal structure of thiamine biosynthesis protein (this)2 from geobacter metallireducens. northeast structural3 genomics consortium target gmr137
|
| 39 | c2dvkA_ |
|
not modelled |
12.2 |
21 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:upf0130 protein ape0816;
PDBTitle: crystal structure of hypothetical protein from aeropyrum pernix
|
| 40 | d1a68a_ |
|
not modelled |
11.9 |
21 |
Fold:POZ domain
Superfamily:POZ domain
Family:Tetramerization domain of potassium channels |
| 41 | d1wria_ |
|
not modelled |
11.3 |
15 |
Fold:beta-Grasp (ubiquitin-like)
Superfamily:2Fe-2S ferredoxin-like
Family:2Fe-2S ferredoxin-related |
| 42 | d1s1ga_ |
|
not modelled |
11.0 |
13 |
Fold:POZ domain
Superfamily:POZ domain
Family:Tetramerization domain of potassium channels |
| 43 | d1st6a4 |
|
not modelled |
10.9 |
24 |
Fold:Four-helical up-and-down bundle
Superfamily:alpha-catenin/vinculin-like
Family:alpha-catenin/vinculin |
| 44 | c3drzE_ |
|
not modelled |
10.9 |
13 |
PDB header:unknown function
Chain: E: PDB Molecule:btb/poz domain-containing protein kctd5;
PDBTitle: x-ray crystal structure of the n-terminal btb domain of human kctd52 protein
|
| 45 | d1frra_ |
|
not modelled |
10.7 |
12 |
Fold:beta-Grasp (ubiquitin-like)
Superfamily:2Fe-2S ferredoxin-like
Family:2Fe-2S ferredoxin-related |
| 46 | c2yy0D_ |
|
not modelled |
10.2 |
12 |
PDB header:transcription
Chain: D: PDB Molecule:c-myc-binding protein;
PDBTitle: crystal structure of ms0802, c-myc-1 binding protein domain2 from homo sapiens
|
| 47 | d3kvta_ |
|
not modelled |
10.0 |
20 |
Fold:POZ domain
Superfamily:POZ domain
Family:Tetramerization domain of potassium channels |
| 48 | c2wukD_ |
|
not modelled |
9.7 |
15 |
PDB header:cell cycle
Chain: D: PDB Molecule:septum site-determining protein diviva;
PDBTitle: diviva n-terminal domain, f17a mutant
|
| 49 | d1i5pa1 |
|
not modelled |
9.6 |
31 |
Fold:Galactose-binding domain-like
Superfamily:Galactose-binding domain-like
Family:delta-Endotoxin, C-terminal domain |
| 50 | d1y6ja2 |
|
not modelled |
9.2 |
26 |
Fold:LDH C-terminal domain-like
Superfamily:LDH C-terminal domain-like
Family:Lactate & malate dehydrogenases, C-terminal domain |
| 51 | d1v86a_ |
|
not modelled |
9.1 |
21 |
Fold:beta-Grasp (ubiquitin-like)
Superfamily:Ubiquitin-like
Family:Ubiquitin-related |
| 52 | c2l5gB_ |
|
not modelled |
9.1 |
16 |
PDB header:transcription regulator
Chain: B: PDB Molecule:putative uncharacterized protein ncor2;
PDBTitle: co-ordinates and 1h, 13c and 15n chemical shift assignments for the2 complex of gps2 53-90 and smrt 167-207
|
| 53 | c1aq5C_ |
|
not modelled |
9.0 |
13 |
PDB header:coiled-coil
Chain: C: PDB Molecule:cartilage matrix protein;
PDBTitle: high-resolution solution nmr structure of the trimeric coiled-coil2 domain of chicken cartilage matrix protein, 20 structures
|
| 54 | c1kd8E_ |
|
not modelled |
8.9 |
10 |
PDB header:de novo protein
Chain: E: PDB Molecule:gcn4 acid base heterodimer base-d12la16l;
PDBTitle: x-ray structure of the coiled coil gcn4 acid base2 heterodimer acid-d12ia16v base-d12la16l
|
| 55 | c3ghgK_ |
|
not modelled |
8.8 |
10 |
PDB header:blood clotting
Chain: K: PDB Molecule:fibrinogen beta chain;
PDBTitle: crystal structure of human fibrinogen
|
| 56 | d1ujra_ |
|
not modelled |
8.8 |
13 |
Fold:WWE domain
Superfamily:WWE domain
Family:WWE domain |
| 57 | c2gzdC_ |
|
not modelled |
8.3 |
16 |
PDB header:protein transport
Chain: C: PDB Molecule:rab11 family-interacting protein 2;
PDBTitle: crystal structure of rab11 in complex with rab11-fip2
|
| 58 | d1lvfa_ |
|
not modelled |
8.2 |
16 |
Fold:STAT-like
Superfamily:t-snare proteins
Family:t-snare proteins |
| 59 | c3iz5J_ |
|
not modelled |
7.9 |
19 |
PDB header:ribosome
Chain: J: PDB Molecule:60s ribosomal protein l12 (l11p);
PDBTitle: localization of the large subunit ribosomal proteins into a 5.5 a2 cryo-em map of triticum aestivum translating 80s ribosome
|
| 60 | c2k4mA_ |
|
not modelled |
7.7 |
19 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:upf0146 protein mth_1000;
PDBTitle: solution nmr structure of m. thermoautotrophicum protein2 mth_1000, northeast structural genomics consortium target3 tr8
|
| 61 | d1hyha2 |
|
not modelled |
7.7 |
19 |
Fold:LDH C-terminal domain-like
Superfamily:LDH C-terminal domain-like
Family:Lactate & malate dehydrogenases, C-terminal domain |
| 62 | d1wqga1 |
|
not modelled |
7.4 |
14 |
Fold:RRF/tRNA synthetase additional domain-like
Superfamily:Ribosome recycling factor, RRF
Family:Ribosome recycling factor, RRF |
| 63 | c2xzrA_ |
|
not modelled |
7.3 |
10 |
PDB header:cell adhesion
Chain: A: PDB Molecule:immunoglobulin-binding protein eibd;
PDBTitle: escherichia coli immunoglobulin-binding protein eibd 391-438 fused2 to gcn4 adaptors
|
| 64 | d2j0111 |
|
not modelled |
7.3 |
10 |
Fold:L28p-like
Superfamily:L28p-like
Family:Ribosomal protein L28 |
| 65 | d1an2a_ |
|
not modelled |
7.2 |
15 |
Fold:HLH-like
Superfamily:HLH, helix-loop-helix DNA-binding domain
Family:HLH, helix-loop-helix DNA-binding domain |
| 66 | c3p8cF_ |
|
not modelled |
7.1 |
18 |
PDB header:protein binding
Chain: F: PDB Molecule:abl interactor 2;
PDBTitle: structure and control of the actin regulatory wave complex
|
| 67 | d1a70a_ |
|
not modelled |
7.1 |
23 |
Fold:beta-Grasp (ubiquitin-like)
Superfamily:2Fe-2S ferredoxin-like
Family:2Fe-2S ferredoxin-related |
| 68 | d1gaqb_ |
|
not modelled |
7.1 |
16 |
Fold:beta-Grasp (ubiquitin-like)
Superfamily:2Fe-2S ferredoxin-like
Family:2Fe-2S ferredoxin-related |
| 69 | d1m94a_ |
|
not modelled |
7.0 |
31 |
Fold:beta-Grasp (ubiquitin-like)
Superfamily:Ubiquitin-like
Family:Ubiquitin-related |
| 70 | c1m94A_ |
|
not modelled |
7.0 |
31 |
PDB header:structural genomics, signaling protein
Chain: A: PDB Molecule:protein ynr032c-a;
PDBTitle: solution structure of the yeast ubiquitin-like modifier2 protein hub1
|
| 71 | d1p0ra_ |
|
not modelled |
6.7 |
38 |
Fold:beta-Grasp (ubiquitin-like)
Superfamily:Ubiquitin-like
Family:Ubiquitin-related |
| 72 | d1qu6a2 |
|
not modelled |
6.7 |
8 |
Fold:dsRBD-like
Superfamily:dsRNA-binding domain-like
Family:Double-stranded RNA-binding domain (dsRBD) |
| 73 | d1jq4a_ |
|
not modelled |
6.7 |
31 |
Fold:beta-Grasp (ubiquitin-like)
Superfamily:2Fe-2S ferredoxin-like
Family:2Fe-2S ferredoxin domains from multidomain proteins |
| 74 | d1dd5a_ |
|
not modelled |
6.6 |
11 |
Fold:RRF/tRNA synthetase additional domain-like
Superfamily:Ribosome recycling factor, RRF
Family:Ribosome recycling factor, RRF |
| 75 | d1i0za2 |
|
not modelled |
6.6 |
26 |
Fold:LDH C-terminal domain-like
Superfamily:LDH C-terminal domain-like
Family:Lactate & malate dehydrogenases, C-terminal domain |
| 76 | d1oqya4 |
|
not modelled |
6.5 |
18 |
Fold:beta-Grasp (ubiquitin-like)
Superfamily:Ubiquitin-like
Family:Ubiquitin-related |
| 77 | c2i2rK_ |
|
not modelled |
6.4 |
13 |
PDB header:transport protein
Chain: K: PDB Molecule:potassium voltage-gated channel subfamily d member 3;
PDBTitle: crystal structure of the kchip1/kv4.3 t1 complex
|
| 78 | d1t3ta3 |
|
not modelled |
6.4 |
19 |
Fold:PurS-like
Superfamily:PurS-like
Family:FGAM synthase PurL, PurS-like domain |
| 79 | c2odkD_ |
|
not modelled |
6.3 |
25 |
PDB header:structural genomics, unknown function
Chain: D: PDB Molecule:hypothetical protein;
PDBTitle: putative prevent-host-death protein from nitrosomonas europaea
|
| 80 | d1nkpa_ |
|
not modelled |
6.0 |
15 |
Fold:HLH-like
Superfamily:HLH, helix-loop-helix DNA-binding domain
Family:HLH, helix-loop-helix DNA-binding domain |
| 81 | c3fp9E_ |
|
not modelled |
5.9 |
39 |
PDB header:hydrolase
Chain: E: PDB Molecule:proteasome-associated atpase;
PDBTitle: crystal structure of intern domain of proteasome-associated2 atpase, mycobacterium tuberculosis
|
| 82 | d1uxja2 |
|
not modelled |
5.9 |
23 |
Fold:LDH C-terminal domain-like
Superfamily:LDH C-terminal domain-like
Family:Lactate & malate dehydrogenases, C-terminal domain |
| 83 | d1awda_ |
|
not modelled |
5.8 |
23 |
Fold:beta-Grasp (ubiquitin-like)
Superfamily:2Fe-2S ferredoxin-like
Family:2Fe-2S ferredoxin-related |
| 84 | d1v97a2 |
|
not modelled |
5.8 |
19 |
Fold:beta-Grasp (ubiquitin-like)
Superfamily:2Fe-2S ferredoxin-like
Family:2Fe-2S ferredoxin domains from multidomain proteins |
| 85 | d2cqla1 |
|
not modelled |
5.8 |
35 |
Fold:Ribosomal protein L6
Superfamily:Ribosomal protein L6
Family:Ribosomal protein L6 |
| 86 | d1whna_ |
|
not modelled |
5.8 |
7 |
Fold:dsRBD-like
Superfamily:dsRNA-binding domain-like
Family:Double-stranded RNA-binding domain (dsRBD) |
| 87 | c2k1iA_ |
|
not modelled |
5.7 |
29 |
PDB header:antimicrobial protein
Chain: A: PDB Molecule:mucosal alpha-defensin;
PDBTitle: synthesis, structure and activities of an oral mucosal2 alpha-defensin from rhesus macaque
|
| 88 | d1gv0a2 |
|
not modelled |
5.7 |
14 |
Fold:LDH C-terminal domain-like
Superfamily:LDH C-terminal domain-like
Family:Lactate & malate dehydrogenases, C-terminal domain |
| 89 | d1nkpb_ |
|
not modelled |
5.6 |
10 |
Fold:HLH-like
Superfamily:HLH, helix-loop-helix DNA-binding domain
Family:HLH, helix-loop-helix DNA-binding domain |
| 90 | c1zmpD_ |
|
not modelled |
5.5 |
42 |
PDB header:antimicrobial protein
Chain: D: PDB Molecule:defensin 5;
PDBTitle: crystal structure of human defensin-5
|
| 91 | d1p9qc2 |
|
not modelled |
5.5 |
17 |
Fold:FYSH domain
Superfamily:FYSH domain
Family:Hypothetical protein AF0491, N-terminal domain |
| 92 | c3m63B_ |
|
not modelled |
5.4 |
19 |
PDB header:ligase/protein binding
Chain: B: PDB Molecule:ubiquitin domain-containing protein dsk2;
PDBTitle: crystal structure of ufd2 in complex with the ubiquitin-like (ubl)2 domain of dsk2
|
| 93 | c3mkxC_ |
|
not modelled |
5.4 |
19 |
PDB header:antiviral protein
Chain: C: PDB Molecule:bone marrow stromal antigen 2;
PDBTitle: crystal structure of bst2/tetherin
|
| 94 | d1krha3 |
|
not modelled |
5.3 |
24 |
Fold:beta-Grasp (ubiquitin-like)
Superfamily:2Fe-2S ferredoxin-like
Family:2Fe-2S ferredoxin domains from multidomain proteins |
| 95 | d2b7ta1 |
|
not modelled |
5.3 |
14 |
Fold:dsRBD-like
Superfamily:dsRNA-binding domain-like
Family:Double-stranded RNA-binding domain (dsRBD) |