| 1 |
|
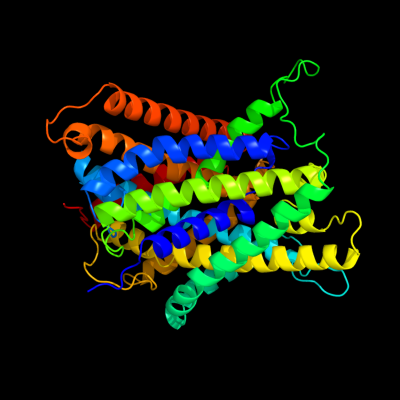
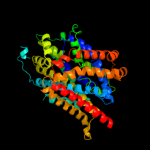
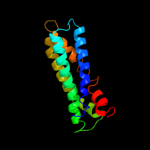
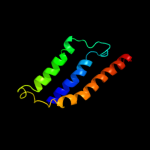
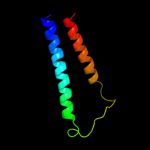
PDB 3gia chain A
Region: 6 - 476
Aligned: 422
Modelled: 422
Confidence: 100.0%
Identity: 13%
PDB header:transport protein
Chain: A: PDB Molecule:uncharacterized protein mj0609;
PDBTitle: crystal structure of apct transporter
Phyre2
| 2 |
|
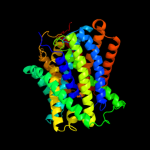
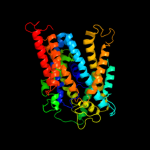
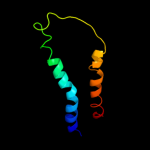
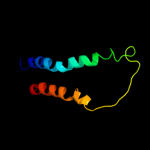
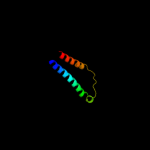
PDB 3lrc chain C
Region: 6 - 476
Aligned: 407
Modelled: 407
Confidence: 100.0%
Identity: 16%
PDB header:transport protein
Chain: C: PDB Molecule:arginine/agmatine antiporter;
PDBTitle: structure of e. coli adic (p1)
Phyre2
| 3 |
|
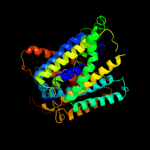
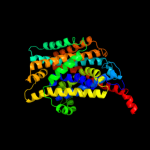
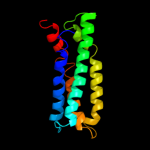
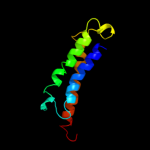
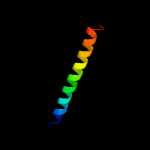
PDB 2jln chain A
Region: 4 - 476
Aligned: 431
Modelled: 431
Confidence: 100.0%
Identity: 9%
PDB header:membrane protein
Chain: A: PDB Molecule:mhp1;
PDBTitle: structure of mhp1, a nucleobase-cation-symport-1 family2 transporter
Phyre2
| 4 |
|
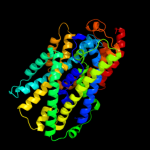
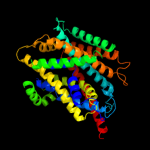
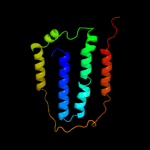
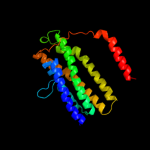
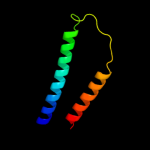
PDB 2a65 chain A domain 1
Region: 11 - 475
Aligned: 452
Modelled: 465
Confidence: 99.2%
Identity: 15%
Fold: SNF-like
Superfamily: SNF-like
Family: SNF-like
Phyre2
| 5 |
|
PDB 2xq2 chain A
Region: 8 - 476
Aligned: 431
Modelled: 453
Confidence: 98.9%
Identity: 10%
PDB header:transport protein
Chain: A: PDB Molecule:sodium/glucose cotransporter;
PDBTitle: structure of the k294a mutant of vsglt
Phyre2
| 6 |
|
PDB 3dh4 chain A
Region: 3 - 476
Aligned: 434
Modelled: 431
Confidence: 98.7%
Identity: 10%
PDB header:transport protein
Chain: A: PDB Molecule:sodium/glucose cotransporter;
PDBTitle: crystal structure of sodium/sugar symporter with bound galactose from2 vibrio parahaemolyticus
Phyre2
| 7 |
|
PDB 2w8a chain C
Region: 3 - 410
Aligned: 393
Modelled: 398
Confidence: 97.8%
Identity: 11%
PDB header:membrane protein
Chain: C: PDB Molecule:glycine betaine transporter betp;
PDBTitle: crystal structure of the sodium-coupled glycine betaine2 symporter betp from corynebacterium glutamicum with bound3 substrate
Phyre2
| 8 |
|
PDB 3hfx chain A
Region: 3 - 402
Aligned: 383
Modelled: 383
Confidence: 92.9%
Identity: 9%
PDB header:transport protein
Chain: A: PDB Molecule:l-carnitine/gamma-butyrobetaine antiporter;
PDBTitle: crystal structure of carnitine transporter
Phyre2
| 9 |
|
PDB 1iwg chain A domain 8
Region: 337 - 476
Aligned: 140
Modelled: 140
Confidence: 70.3%
Identity: 12%
Fold: Multidrug efflux transporter AcrB transmembrane domain
Superfamily: Multidrug efflux transporter AcrB transmembrane domain
Family: Multidrug efflux transporter AcrB transmembrane domain
Phyre2
| 10 |
|
PDB 1fft chain B domain 2
Region: 367 - 434
Aligned: 68
Modelled: 68
Confidence: 60.2%
Identity: 10%
Fold: Transmembrane helix hairpin
Superfamily: Cytochrome c oxidase subunit II-like, transmembrane region
Family: Cytochrome c oxidase subunit II-like, transmembrane region
Phyre2
| 11 |
|
PDB 1oy8 chain A
Region: 337 - 476
Aligned: 140
Modelled: 140
Confidence: 56.3%
Identity: 11%
PDB header:membrane protein
Chain: A: PDB Molecule:acriflavine resistance protein b;
PDBTitle: structural basis of multiple drug binding capacity of the acrb2 multidrug efflux pump
Phyre2
| 12 |
|
PDB 3rko chain F
Region: 338 - 475
Aligned: 131
Modelled: 138
Confidence: 36.0%
Identity: 8%
PDB header:oxidoreductase
Chain: F: PDB Molecule:nadh-quinone oxidoreductase subunit j;
PDBTitle: crystal structure of the membrane domain of respiratory complex i from2 e. coli at 3.0 angstrom resolution
Phyre2
| 13 |
|
PDB 3b8e chain C
Region: 380 - 476
Aligned: 97
Modelled: 97
Confidence: 21.0%
Identity: 9%
PDB header:hydrolase/transport protein
Chain: C: PDB Molecule:sodium/potassium-transporting atpase subunit
PDBTitle: crystal structure of the sodium-potassium pump
Phyre2
| 14 |
|
PDB 1fft chain G
Region: 366 - 434
Aligned: 69
Modelled: 69
Confidence: 16.8%
Identity: 10%
PDB header:oxidoreductase
Chain: G: PDB Molecule:ubiquinol oxidase;
PDBTitle: the structure of ubiquinol oxidase from escherichia coli
Phyre2
| 15 |
|
PDB 3ixz chain A
Region: 380 - 476
Aligned: 97
Modelled: 97
Confidence: 11.0%
Identity: 8%
PDB header:hydrolase
Chain: A: PDB Molecule:potassium-transporting atpase alpha;
PDBTitle: pig gastric h+/k+-atpase complexed with aluminium fluoride
Phyre2
| 16 |
|
PDB 3aqp chain B
Region: 229 - 435
Aligned: 197
Modelled: 207
Confidence: 10.3%
Identity: 9%
PDB header:membrane protein
Chain: B: PDB Molecule:probable secdf protein-export membrane protein;
PDBTitle: crystal structure of secdf, a translocon-associated membrane protein,2 from thermus thrmophilus
Phyre2
| 17 |
|
PDB 1m57 chain H
Region: 366 - 434
Aligned: 67
Modelled: 69
Confidence: 9.7%
Identity: 13%
PDB header:oxidoreductase
Chain: H: PDB Molecule:cytochrome c oxidase;
PDBTitle: structure of cytochrome c oxidase from rhodobacter2 sphaeroides (eq(i-286) mutant))
Phyre2
| 18 |
|
PDB 3ehb chain B domain 2
Region: 366 - 429
Aligned: 64
Modelled: 64
Confidence: 8.1%
Identity: 13%
Fold: Transmembrane helix hairpin
Superfamily: Cytochrome c oxidase subunit II-like, transmembrane region
Family: Cytochrome c oxidase subunit II-like, transmembrane region
Phyre2
| 19 |
|
PDB 2jwa chain A
Region: 443 - 476
Aligned: 34
Modelled: 34
Confidence: 8.0%
Identity: 24%
PDB header:transferase
Chain: A: PDB Molecule:receptor tyrosine-protein kinase erbb-2;
PDBTitle: erbb2 transmembrane segment dimer spatial structure
Phyre2
| 20 |
|
PDB 3dtu chain B domain 2
Region: 366 - 429
Aligned: 64
Modelled: 64
Confidence: 7.5%
Identity: 13%
Fold: Transmembrane helix hairpin
Superfamily: Cytochrome c oxidase subunit II-like, transmembrane region
Family: Cytochrome c oxidase subunit II-like, transmembrane region
Phyre2
| 21 |
|
| 22 |
|