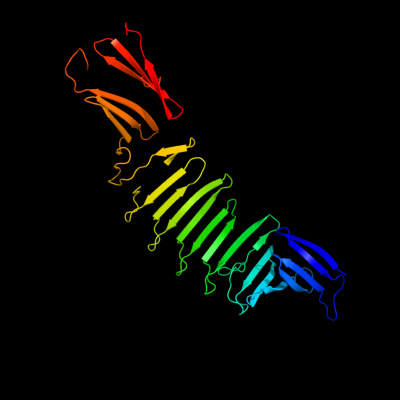
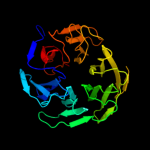
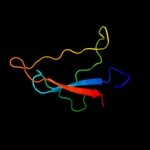
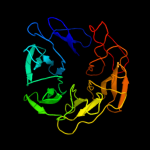
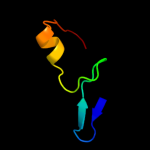
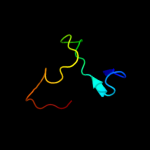
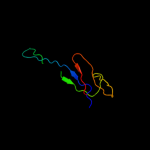
1 c2oy7A_
96.5
15
PDB header: membrane proteinChain: A: PDB Molecule: outer surface protein a;PDBTitle: the crystal structure of ospa mutant
2 d1hnga1
92.7
19
Fold: Immunoglobulin-like beta-sandwichSuperfamily: ImmunoglobulinFamily: V set domains (antibody variable domain-like)3 d1hnfa1
68.3
16
Fold: Immunoglobulin-like beta-sandwichSuperfamily: ImmunoglobulinFamily: V set domains (antibody variable domain-like)4 c1n7dA_
67.7
12
PDB header: lipid transportChain: A: PDB Molecule: low-density lipoprotein receptor;PDBTitle: extracellular domain of the ldl receptor
5 d1x3za1
39.1
18
Fold: Cysteine proteinasesSuperfamily: Cysteine proteinasesFamily: Transglutaminase core6 d1ospo_
33.3
16
Fold: open-sided beta-meanderSuperfamily: Outer surface proteinFamily: Outer surface protein7 c2w8bB_
31.4
12
PDB header: protein transport/membrane proteinChain: B: PDB Molecule: protein tolb;PDBTitle: crystal structure of processed tolb in complex with pal
8 c3e5zA_
31.0
15
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: putative gluconolactonase;PDBTitle: x-ray structure of the putative gluconolactonase in protein family2 pf08450. northeast structural genomics consortium target drr130.
9 c3rwxA_
25.0
18
PDB header: transport proteinChain: A: PDB Molecule: hypothetical bacterial outer membrane protein;PDBTitle: crystal structure of a hypothetical bacterial outer membrane protein2 (bf2706) from bacteroides fragilis nctc 9343 at 2.40 a resolution
10 d2j44a2
23.8
18
Fold: Prealbumin-likeSuperfamily: Starch-binding domain-likeFamily: PUD-like11 c2ivzD_
22.0
11
PDB header: protein transport/hydrolaseChain: D: PDB Molecule: protein tolb;PDBTitle: structure of tolb in complex with a peptide of the colicin2 e9 t-domain
12 d2f4ma1
21.0
26
Fold: Cysteine proteinasesSuperfamily: Cysteine proteinasesFamily: Transglutaminase core13 c3eswA_
20.8
21
PDB header: hydrolaseChain: A: PDB Molecule: peptide-n(4)-(n-acetyl-beta-glucosaminyl)asparaginePDBTitle: complex of yeast pngase with glcnac2-iac.
14 d1sska_
20.3
21
Fold: Coronavirus RNA-binding domainSuperfamily: Coronavirus RNA-binding domainFamily: Coronavirus RNA-binding domain15 c3mswA_
20.3
20
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: uncharacterized protein;PDBTitle: crystal structure of a protein with unknown function (bf3112) from2 bacteroides fragilis nctc 9343 at 1.90 a resolution
16 c1j5qB_
20.0
28
PDB header: viral proteinChain: B: PDB Molecule: major capsid protein;PDBTitle: the structure and evolution of the major capsid protein of a large,2 lipid-containing, dna virus.
17 d2j43a1
17.2
16
Fold: Prealbumin-likeSuperfamily: Starch-binding domain-likeFamily: PUD-like18 d2j44a1
17.2
15
Fold: Prealbumin-likeSuperfamily: Starch-binding domain-likeFamily: PUD-like19 d1ei5a1
15.5
32
Fold: Streptavidin-likeSuperfamily: D-aminopeptidase, middle and C-terminal domainsFamily: D-aminopeptidase, middle and C-terminal domains20 c3k8hA_
15.5
24
PDB header: membrane proteinChain: A: PDB Molecule: 30klp;PDBTitle: structure of crystal form i of tp0453
21 c1t3dB_
not modelled
14.0
32
PDB header: transferaseChain: B: PDB Molecule: serine acetyltransferase;PDBTitle: crystal structure of serine acetyltransferase from e.coli at 2.2a
22 c3hd4A_
not modelled
13.8
21
PDB header: viral proteinChain: A: PDB Molecule: nucleoprotein;PDBTitle: mhv nucleocapsid protein ntd
23 c3ilwA_
not modelled
13.7
24
PDB header: isomeraseChain: A: PDB Molecule: dna gyrase subunit a;PDBTitle: structure of dna gyrase subunit a n-terminal domain
24 d1o5ua_
not modelled
13.6
8
Fold: Double-stranded beta-helixSuperfamily: RmlC-like cupinsFamily: Hypothetical protein TM111225 c2ksnA_
not modelled
13.1
20
PDB header: signaling proteinChain: A: PDB Molecule: ubiquitin domain-containing protein 2;PDBTitle: solution structure of the n-terminal domain of dc-ubp/ubtd2
26 c2x12A_
not modelled
12.6
27
PDB header: cell adhesionChain: A: PDB Molecule: fimbriae-associated protein fap1;PDBTitle: ph-induced modulation of streptococcus parasanguinis2 adhesion by fap1 fimbriae
27 c3s88I_
not modelled
12.6
19
PDB header: immune system/viral proteinChain: I: PDB Molecule: envelope glycoprotein;PDBTitle: crystal structure of sudan ebolavirus glycoprotein (strain gulu) bound2 to 16f6
28 c2kijA_
not modelled
12.1
25
PDB header: hydrolaseChain: A: PDB Molecule: copper-transporting atpase 1;PDBTitle: solution structure of the actuator domain of the copper-2 transporting atpase atp7a
29 c3bmxB_
not modelled
12.1
42
PDB header: hydrolaseChain: B: PDB Molecule: uncharacterized lipoprotein ybbd;PDBTitle: beta-n-hexosaminidase (ybbd) from bacillus subtilis
30 d1xata_
not modelled
11.8
26
Fold: Single-stranded left-handed beta-helixSuperfamily: Trimeric LpxA-like enzymesFamily: Galactoside acetyltransferase-like31 d1t3da_
not modelled
11.6
32
Fold: Single-stranded left-handed beta-helixSuperfamily: Trimeric LpxA-like enzymesFamily: Serine acetyltransferase32 d1pk6c_
not modelled
11.4
27
Fold: TNF-likeSuperfamily: TNF-likeFamily: TNF-like33 c2wlgA_
not modelled
11.2
26
PDB header: transferaseChain: A: PDB Molecule: polysialic acid o-acetyltransferase;PDBTitle: crystallographic analysis of the polysialic acid o-2 acetyltransferase oatwy
34 d1td6a_
not modelled
11.1
31
Fold: Hypothetical protein MPN330Superfamily: Hypothetical protein MPN330Family: Hypothetical protein MPN33035 c2j44A_
not modelled
11.1
15
PDB header: carbohydrate-binding moduleChain: A: PDB Molecule: alkaline amylopullulanase;PDBTitle: alpha-glucan binding by a streptococcal virulence factor
36 c2gopB_
not modelled
11.0
8
PDB header: hydrolaseChain: B: PDB Molecule: trilobed protease;PDBTitle: the beta-propeller domain of the trilobed protease from pyrococcus2 furiosus reveals an open velcro topology
37 d1bcoa1
not modelled
11.0
17
Fold: mu transposase, C-terminal domainSuperfamily: mu transposase, C-terminal domainFamily: mu transposase, C-terminal domain38 c3ifzA_
not modelled
10.8
21
PDB header: isomeraseChain: A: PDB Molecule: dna gyrase subunit a;PDBTitle: crystal structure of the first part of the mycobacterium tuberculosis2 dna gyrase reaction core: the breakage and reunion domain at 2.7 a3 resolution
39 d2bxxa1
not modelled
10.8
24
Fold: Coronavirus RNA-binding domainSuperfamily: Coronavirus RNA-binding domainFamily: Coronavirus RNA-binding domain40 c2ka3C_
not modelled
10.7
36
PDB header: structural proteinChain: C: PDB Molecule: emilin-1;PDBTitle: structure of emilin-1 c1q-like domain
41 d1brwa3
not modelled
10.5
26
Fold: alpha/beta-HammerheadSuperfamily: Pyrimidine nucleoside phosphorylase C-terminal domainFamily: Pyrimidine nucleoside phosphorylase C-terminal domain42 d2i52a1
not modelled
10.2
17
Fold: MK0786-likeSuperfamily: MK0786-likeFamily: MK0786-like43 d1bcga_
not modelled
10.2
38
Fold: Knottins (small inhibitors, toxins, lectins)Superfamily: Scorpion toxin-likeFamily: Long-chain scorpion toxins44 d1pk6b_
not modelled
10.1
27
Fold: TNF-likeSuperfamily: TNF-likeFamily: TNF-like45 d1pk6a_
not modelled
10.1
27
Fold: TNF-likeSuperfamily: TNF-likeFamily: TNF-like46 c3jroA_
not modelled
10.0
12
PDB header: transport protein, structural proteinChain: A: PDB Molecule: fusion protein of protein transport protein sec13PDBTitle: nup84-nup145c-sec13 edge element of the npc lattice
47 d1mr7a_
not modelled
9.7
24
Fold: Single-stranded left-handed beta-helixSuperfamily: Trimeric LpxA-like enzymesFamily: Galactoside acetyltransferase-like48 d2tpta3
not modelled
9.7
23
Fold: alpha/beta-HammerheadSuperfamily: Pyrimidine nucleoside phosphorylase C-terminal domainFamily: Pyrimidine nucleoside phosphorylase C-terminal domain49 d2jdqd1
not modelled
9.5
20
Fold: PB2 C-terminal domain-likeSuperfamily: PB2 C-terminal domain-likeFamily: PB2 C-terminal domain-like50 d1f00i2
not modelled
9.5
38
Fold: Immunoglobulin-like beta-sandwichSuperfamily: Invasin/intimin cell-adhesion fragmentsFamily: Invasin/intimin cell-adhesion fragments51 d1m3ya2
not modelled
9.4
39
Fold: Nucleoplasmin-like/VP (viral coat and capsid proteins)Superfamily: Group II dsDNA viruses VPFamily: Major capsid protein vp5452 c3bgyA_
not modelled
9.4
20
PDB header: hydrolase, viral proteinChain: A: PDB Molecule: polynucleotide 5'-triphosphatase;PDBTitle: triclinic structure of mimivirus capping enzyme2 triphosphatase at 1.65 a
53 c3fsbB_
not modelled
9.1
27
PDB header: transferaseChain: B: PDB Molecule: qdtc;PDBTitle: crystal structure of qdtc, the dtdp-3-amino-3,6-dideoxy-d-2 glucose n-acetyl transferase from thermoanaerobacterium3 thermosaccharolyticum in complex with coa and dtdp-3-amino-4 quinovose
54 d1c3ha_
not modelled
9.0
31
Fold: TNF-likeSuperfamily: TNF-likeFamily: TNF-like55 c3vh0C_
not modelled
9.0
9
PDB header: protein binding/dnaChain: C: PDB Molecule: uncharacterized protein ynce;PDBTitle: crystal structure of e. coli ynce complexed with dna
56 c2hc8A_
not modelled
9.0
28
PDB header: transport proteinChain: A: PDB Molecule: cation-transporting atpase, p-type;PDBTitle: structure of the a. fulgidus copa a-domain
57 d1krra_
not modelled
8.9
29
Fold: Single-stranded left-handed beta-helixSuperfamily: Trimeric LpxA-like enzymesFamily: Galactoside acetyltransferase-like58 c1gr3A_
not modelled
8.9
45
PDB header: collagenChain: A: PDB Molecule: collagen x;PDBTitle: structure of the human collagen x nc1 trimer
59 d1gr3a_
not modelled
8.9
45
Fold: TNF-likeSuperfamily: TNF-likeFamily: TNF-like60 d1j2za_
not modelled
8.7
19
Fold: Single-stranded left-handed beta-helixSuperfamily: Trimeric LpxA-like enzymesFamily: UDP N-acetylglucosamine acyltransferase61 c2xzmG_
not modelled
8.7
17
PDB header: ribosomeChain: G: PDB Molecule: ribosomal protein s7 containing protein;PDBTitle: crystal structure of the eukaryotic 40s ribosomal2 subunit in complex with initiation factor 1. this file3 contains the 40s subunit and initiation factor for4 molecule 1
62 c2iu9C_
not modelled
8.6
23
PDB header: transferaseChain: C: PDB Molecule: udp-3-o-[3-hydroxymyristoyl] glucosaminePDBTitle: chlamydia trachomatis lpxd with 100mm udpglcnac (complex ii)
63 d1jnpa_
not modelled
8.6
26
Fold: Oncogene productsSuperfamily: Oncogene productsFamily: Oncogene products64 c1m4xC_
not modelled
8.5
39
PDB header: virusChain: C: PDB Molecule: pbcv-1 virus capsid;PDBTitle: pbcv-1 virus capsid, quasi-atomic model
65 c2vtwF_
not modelled
8.4
32
PDB header: viral proteinChain: F: PDB Molecule: fiber protein 2;PDBTitle: structure of the c-terminal head domain of the fowl2 adenovirus type 1 short fibre
66 d2geca1
not modelled
8.3
24
Fold: Coronavirus RNA-binding domainSuperfamily: Coronavirus RNA-binding domainFamily: Coronavirus RNA-binding domain67 c3di4A_
not modelled
8.3
25
PDB header: metal binding proteinChain: A: PDB Molecule: uncharacterized protein duf1989;PDBTitle: crystal structure of a duf1989 family protein (spo0365) from2 silicibacter pomeroyi dss-3 at 1.60 a resolution
68 d1azpa_
not modelled
7.9
17
Fold: SH3-like barrelSuperfamily: Chromo domain-likeFamily: "Histone-like" proteins from archaea69 d1a1xa_
not modelled
7.9
21
Fold: Oncogene productsSuperfamily: Oncogene productsFamily: Oncogene products70 c3qx3B_
not modelled
7.9
34
PDB header: isomerase/dna/isomerase inhibitorChain: B: PDB Molecule: dna topoisomerase 2-beta;PDBTitle: human topoisomerase iibeta in complex with dna and etoposide
71 d1ufya_
not modelled
7.8
13
Fold: Bacillus chorismate mutase-likeSuperfamily: YjgF-likeFamily: Chorismate mutase72 d1f1sa3
not modelled
7.8
18
Fold: Hyaluronate lyase-like, C-terminal domainSuperfamily: Hyaluronate lyase-like, C-terminal domainFamily: Hyaluronate lyase-like, C-terminal domain73 c3eevC_
not modelled
7.8
26
PDB header: transferaseChain: C: PDB Molecule: chloramphenicol acetyltransferase;PDBTitle: crystal structure of chloramphenicol acetyltransferase vca0300 from2 vibrio cholerae o1 biovar eltor
74 d2oqea2
not modelled
7.6
25
Fold: Cystatin-likeSuperfamily: Amine oxidase N-terminal regionFamily: Amine oxidase N-terminal region75 d2pyta1
not modelled
7.5
13
Fold: Double-stranded beta-helixSuperfamily: RmlC-like cupinsFamily: EutQ-like76 c3myxA_
not modelled
7.5
16
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: uncharacterized protein pspto_0244;PDBTitle: crystal structure of a pspto_0244 (protein with unknown function which2 belongs to pfam duf861 family) from pseudomonas syringae pv. tomato3 str. dc3000 at 1.30 a resolution
77 c3r0sA_
not modelled
7.4
18
PDB header: transferaseChain: A: PDB Molecule: acyl-[acyl-carrier-protein]--udp-n-acetylglucosamine o-PDBTitle: udp-n-acetylglucosamine acyltransferase from campylobacter jejuni
78 c2kssA_
not modelled
7.3
31
PDB header: transcription regulatorChain: A: PDB Molecule: carotenogenesis protein cars;PDBTitle: nmr structure of myxococcus xanthus antirepressor cars1
79 c2gacA_
not modelled
7.1
13
PDB header: hydrolaseChain: A: PDB Molecule: glycosylasparaginase;PDBTitle: t152c mutant glycosylasparaginase from flavobacterium2 meningosepticum
80 d1jsga_
not modelled
7.1
26
Fold: Oncogene productsSuperfamily: Oncogene productsFamily: Oncogene products81 d1k8wa3
not modelled
7.0
27
Fold: PUA domain-likeSuperfamily: PUA domain-likeFamily: PUA domain82 d1w9pa2
not modelled
7.0
18
Fold: FKBP-likeSuperfamily: Chitinase insertion domainFamily: Chitinase insertion domain83 c2j43A_
not modelled
7.0
16
PDB header: carbohydrate-binding moduleChain: A: PDB Molecule: spydx;PDBTitle: alpha-glucan recognition by family 41 carbohydrate-binding2 modules from streptococcal virulence factors
84 d1jofa_
not modelled
7.0
13
Fold: 7-bladed beta-propellerSuperfamily: 3-carboxy-cis,cis-mucoante lactonizing enzymeFamily: 3-carboxy-cis,cis-mucoante lactonizing enzyme85 c2l92A_
not modelled
6.9
44
PDB header: dna binding proteinChain: A: PDB Molecule: histone family protein nucleoid-structuring protein h-ns;PDBTitle: solution structure of the c-terminal domain of h-ns like protein bv3f
86 d1oi0a_
not modelled
6.9
15
Fold: Cytidine deaminase-likeSuperfamily: JAB1/MPN domainFamily: JAB1/MPN domain87 c7apiB_
not modelled
6.9
6
PDB header: proteinase inhibitorChain: B: PDB Molecule: alpha 1-antitrypsin;PDBTitle: the s variant of human alpha1-antitrypsin, structure and implications2 for function and metabolism
88 c3i3aC_
not modelled
6.8
19
PDB header: transferaseChain: C: PDB Molecule: acyl-[acyl-carrier-protein]--udp-n-PDBTitle: structural basis for the sugar nucleotide and acyl chain2 selectivity of leptospira interrogans lpxa
89 d2ax3a2
not modelled
6.8
14
Fold: YjeF N-terminal domain-likeSuperfamily: YjeF N-terminal domain-likeFamily: YjeF N-terminal domain-like90 c1jjoE_
not modelled
6.8
22
PDB header: signaling proteinChain: E: PDB Molecule: neuroserpin;PDBTitle: crystal structure of mouse neuroserpin (cleaved form)
91 d1jiwi_
not modelled
6.8
15
Fold: Streptavidin-likeSuperfamily: beta-Barrel protease inhibitorsFamily: Metalloprotease inhibitor92 c1n0nB_
not modelled
6.7
57
PDB header: oxidoreductaseChain: B: PDB Molecule: superoxide dismutase [mn];PDBTitle: catalytic and structural effects of amino-acid substitution at his302 in human manganese superoxide dismutase
93 c3jqyB_
not modelled
6.7
30
PDB header: transferaseChain: B: PDB Molecule: polysialic acid o-acetyltransferase;PDBTitle: crystal strucutre of the polysia specific acetyltransferase neuo
94 c3izbF_
not modelled
6.7
14
PDB header: ribosomeChain: F: PDB Molecule: 40s ribosomal protein rps5 (s7p);PDBTitle: localization of the small subunit ribosomal proteins into a 6.1 a2 cryo-em map of saccharomyces cerevisiae translating 80s ribosome
95 c1y10C_
not modelled
6.7
32
PDB header: lyaseChain: C: PDB Molecule: hypothetical protein rv1264/mt1302;PDBTitle: mycobacterial adenylyl cyclase rv1264, holoenzyme, inhibited state
96 c1p7gL_
not modelled
6.6
43
PDB header: oxidoreductaseChain: L: PDB Molecule: superoxide dismutase;PDBTitle: crystal structure of superoxide dismutase from pyrobaculum2 aerophilum
97 d1ulva3
not modelled
6.6
35
Fold: Immunoglobulin-like beta-sandwichSuperfamily: CBD9-likeFamily: Glucodextranase, domain C98 c1gn4B_
not modelled
6.6
43
PDB header: oxidoreductaseChain: B: PDB Molecule: superoxide dismutase;PDBTitle: h145e mutant of mycobacterium tuberculosis iron-superoxide2 dismutase.
99 c3dr2A_
not modelled
6.6
15
PDB header: hydrolaseChain: A: PDB Molecule: exported gluconolactonase;PDBTitle: structural and functional analyses of xc5397 from2 xanthomonas campestris: a gluconolactonase important in3 glucose secondary metabolic pathways