| 1 |
|
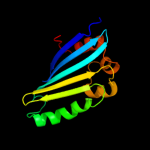
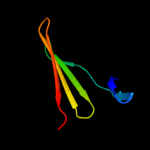
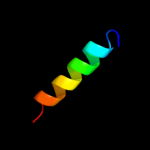
PDB 2joe chain A domain 1
Region: 24 - 153
Aligned: 130
Modelled: 130
Confidence: 100.0%
Identity: 100%
Fold: YehR-like
Superfamily: YehR-like
Family: YehR-like
Phyre2
| 2 |
|
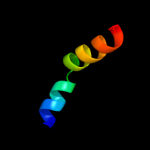
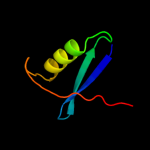
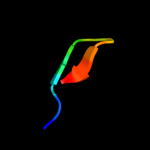
PDB 2duy chain A domain 1
Region: 108 - 125
Aligned: 18
Modelled: 18
Confidence: 19.2%
Identity: 17%
Fold: SAM domain-like
Superfamily: RuvA domain 2-like
Family: ComEA-like
Phyre2
| 3 |
|
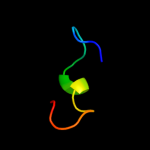
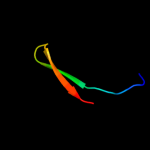
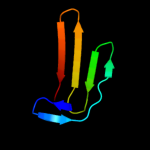
PDB 1jb0 chain J
Region: 1 - 24
Aligned: 24
Modelled: 24
Confidence: 18.9%
Identity: 21%
Fold: Single transmembrane helix
Superfamily: Subunit IX of photosystem I reaction centre, PsaJ
Family: Subunit IX of photosystem I reaction centre, PsaJ
Phyre2
| 4 |
|
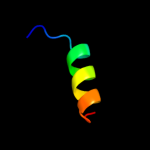
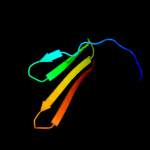
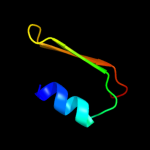
PDB 2axt chain U domain 1
Region: 108 - 125
Aligned: 18
Modelled: 18
Confidence: 15.4%
Identity: 22%
Fold: SAM domain-like
Superfamily: PsbU/PolX domain-like
Family: PsbU-like
Phyre2
| 5 |
|
PDB 1s5l chain U
Region: 108 - 125
Aligned: 18
Modelled: 18
Confidence: 12.6%
Identity: 22%
PDB header:photosynthesis
Chain: U: PDB Molecule:photosystem ii 12 kda extrinsic protein;
PDBTitle: architecture of the photosynthetic oxygen evolving center
Phyre2
| 6 |
|
PDB 2fi0 chain A domain 1
Region: 133 - 150
Aligned: 18
Modelled: 18
Confidence: 12.0%
Identity: 22%
Fold: SP0561-like
Superfamily: SP0561-like
Family: SP0561-like
Phyre2
| 7 |
|
PDB 1lfo chain A
Region: 32 - 109
Aligned: 47
Modelled: 47
Confidence: 10.5%
Identity: 13%
Fold: Lipocalins
Superfamily: Lipocalins
Family: Fatty acid binding protein-like
Phyre2
| 8 |
|
PDB 1wmh chain A
Region: 40 - 100
Aligned: 49
Modelled: 50
Confidence: 10.4%
Identity: 10%
Fold: beta-Grasp (ubiquitin-like)
Superfamily: CAD & PB1 domains
Family: PB1 domain
Phyre2
| 9 |
|
PDB 1p6p chain A
Region: 36 - 109
Aligned: 42
Modelled: 42
Confidence: 9.9%
Identity: 24%
Fold: Lipocalins
Superfamily: Lipocalins
Family: Fatty acid binding protein-like
Phyre2
| 10 |
|
PDB 1zoq chain A
Region: 40 - 59
Aligned: 20
Modelled: 20
Confidence: 8.9%
Identity: 30%
PDB header:transcription/transferase
Chain: A: PDB Molecule:interferon regulatory factor 3;
PDBTitle: irf3-cbp complex
Phyre2
| 11 |
|
PDB 3dsh chain A
Region: 40 - 59
Aligned: 20
Modelled: 20
Confidence: 8.4%
Identity: 15%
PDB header:dna binding protein
Chain: A: PDB Molecule:interferon regulatory factor 5;
PDBTitle: crystal structure of dimeric interferon regulatory factor 5 (irf-5)2 transactivation domain
Phyre2
| 12 |
|
PDB 3pn1 chain A
Region: 88 - 107
Aligned: 20
Modelled: 20
Confidence: 8.4%
Identity: 15%
PDB header:ligase/ligase inhibitor
Chain: A: PDB Molecule:dna ligase;
PDBTitle: novel bacterial nad+-dependent dna ligase inhibitors with broad2 spectrum potency and antibacterial efficacy in vivo
Phyre2
| 13 |
|
PDB 2dzl chain A
Region: 13 - 30
Aligned: 18
Modelled: 18
Confidence: 8.0%
Identity: 22%
PDB header:structural genomics unknown function
Chain: A: PDB Molecule:protein fam100b;
PDBTitle: solution structure of the uba domain in human protein2 fam100b
Phyre2
| 14 |
|
PDB 3em0 chain A
Region: 37 - 109
Aligned: 41
Modelled: 41
Confidence: 8.0%
Identity: 15%
PDB header:lipid binding protein
Chain: A: PDB Molecule:ileal bile acid-binding protein;
PDBTitle: crystal structure of zebrafish ileal bile acid-bindin protein2 complexed with cholic acid (crystal form b).
Phyre2
| 15 |
|
PDB 1im8 chain A
Region: 135 - 153
Aligned: 19
Modelled: 19
Confidence: 7.1%
Identity: 21%
Fold: S-adenosyl-L-methionine-dependent methyltransferases
Superfamily: S-adenosyl-L-methionine-dependent methyltransferases
Family: Hypothetical protein HI0319 (YecO)
Phyre2
| 16 |
|
PDB 1qwt chain A
Region: 40 - 59
Aligned: 20
Modelled: 20
Confidence: 6.2%
Identity: 30%
Fold: SMAD/FHA domain
Superfamily: SMAD/FHA domain
Family: Interferon regulatory factor 3 (IRF3), transactivation domain
Phyre2
| 17 |
|
PDB 3hkz chain Y
Region: 67 - 85
Aligned: 19
Modelled: 19
Confidence: 6.2%
Identity: 37%
PDB header:transferase
Chain: Y: PDB Molecule:dna-directed rna polymerase subunit 13;
PDBTitle: the x-ray crystal structure of rna polymerase from archaea
Phyre2
| 18 |
|
PDB 3kz5 chain E
Region: 44 - 53
Aligned: 10
Modelled: 10
Confidence: 5.9%
Identity: 60%
PDB header:dna binding protein
Chain: E: PDB Molecule:protein sopb;
PDBTitle: structure of cdomain
Phyre2
| 19 |
|
PDB 2ftb chain A domain 1
Region: 33 - 109
Aligned: 45
Modelled: 45
Confidence: 5.8%
Identity: 18%
Fold: Lipocalins
Superfamily: Lipocalins
Family: Fatty acid binding protein-like
Phyre2
| 20 |
|
PDB 1fe0 chain A
Region: 79 - 109
Aligned: 31
Modelled: 31
Confidence: 5.7%
Identity: 13%
Fold: Ferredoxin-like
Superfamily: HMA, heavy metal-associated domain
Family: HMA, heavy metal-associated domain
Phyre2
| 21 |
|
| 22 |
|