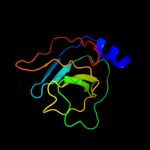
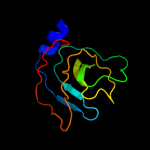
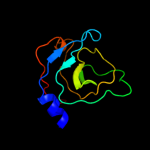
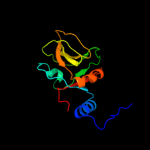
1 c3mmlE_
100.0
35
PDB header: hydrolaseChain: E: PDB Molecule: allophanate hydrolase subunit 2;PDBTitle: allophanate hydrolase complex from mycobacterium smegmatis, msmeg0435-2 msmeg0436
2 c3oepA_
100.0
35
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: putative uncharacterized protein ttha0988;PDBTitle: crystal structure of ttha0988 in space group p43212
3 c2qf7A_
97.3
8
PDB header: ligaseChain: A: PDB Molecule: pyruvate carboxylase protein;PDBTitle: crystal structure of a complete multifunctional pyruvate carboxylase2 from rhizobium etli
4 c2phcB_
96.1
15
PDB header: structural genomics, unknown functionChain: B: PDB Molecule: uncharacterized protein ph0987;PDBTitle: crystal structure of conserved uncharacterized protein ph0987 from2 pyrococcus horikoshii
5 d2phcb1
95.6
15
Fold: Cyclophilin-likeSuperfamily: Cyclophilin-likeFamily: PH0987 C-terminal domain-like6 c2zp2B_
95.4
16
PDB header: transferase inhibitorChain: B: PDB Molecule: kinase a inhibitor;PDBTitle: c-terminal domain of kipi from bacillus subtilis
7 c2p0oA_
93.8
16
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: hypothetical protein duf871;PDBTitle: crystal structure of a conserved protein from locus ef_2437 in2 enterococcus faecalis with an unknown function
8 c1x7fA_
91.9
14
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: outer surface protein;PDBTitle: crystal structure of an uncharacterized b. cereus protein
9 d1x7fa1
91.9
13
Fold: Cyclophilin-likeSuperfamily: Cyclophilin-likeFamily: Outer surface protein, C-terminal domain10 c3mmlD_
89.6
15
PDB header: hydrolaseChain: D: PDB Molecule: allophanate hydrolase subunit 1;PDBTitle: allophanate hydrolase complex from mycobacterium smegmatis, msmeg0435-2 msmeg0436
11 c3elsA_
86.3
17
PDB header: splicingChain: A: PDB Molecule: pre-mrna leakage protein 1;PDBTitle: crystal structure of yeast pml1p, residues 51-204
12 d2piea1
84.9
10
Fold: SMAD/FHA domainSuperfamily: SMAD/FHA domainFamily: FHA domain13 c1gxcA_
81.4
15
PDB header: phosphoprotein-binding domainChain: A: PDB Molecule: serine/threonine-protein kinase chk2;PDBTitle: fha domain from human chk2 kinase in complex with a2 synthetic phosphopeptide
14 d1gxca_
81.4
15
Fold: SMAD/FHA domainSuperfamily: SMAD/FHA domainFamily: FHA domain15 c2jkdB_
80.1
10
PDB header: gene regulationChain: B: PDB Molecule: pre-mrna leakage protein 1;PDBTitle: structure of the yeast pml1 splicing factor and its2 integration into the res complex
16 d1uhta_
69.2
19
Fold: SMAD/FHA domainSuperfamily: SMAD/FHA domainFamily: FHA domain17 c2jpeA_
68.0
2
PDB header: transcriptionChain: A: PDB Molecule: nuclear inhibitor of protein phosphatase 1;PDBTitle: fha domain of nipp1
18 c3gmgB_
67.8
21
PDB header: structural genomics, unknown functionChain: B: PDB Molecule: uncharacterized protein rv1825/mt1873;PDBTitle: crystal structure of an uncharacterized conserved protein2 from mycobacterium tuberculosis
19 d2brfa1
66.5
13
Fold: SMAD/FHA domainSuperfamily: SMAD/FHA domainFamily: FHA domain20 d1ujxa_
65.8
7
Fold: SMAD/FHA domainSuperfamily: SMAD/FHA domainFamily: FHA domain21 c1yj5C_
not modelled
62.8
11
PDB header: transferaseChain: C: PDB Molecule: 5' polynucleotide kinase-3' phosphatase fha domain;PDBTitle: molecular architecture of mammalian polynucleotide kinase, a dna2 repair enzyme
22 c3poaA_
not modelled
62.3
10
PDB header: peptide binding proteinChain: A: PDB Molecule: putative uncharacterized protein tb39.8;PDBTitle: structural and functional analysis of phosphothreonine-dependent fha2 domain interactions
23 c3kt9A_
not modelled
61.7
16
PDB header: hydrolaseChain: A: PDB Molecule: aprataxin;PDBTitle: aprataxin fha domain
24 d1g3ga_
not modelled
61.5
16
Fold: SMAD/FHA domainSuperfamily: SMAD/FHA domainFamily: FHA domain25 d2g1la1
not modelled
54.2
20
Fold: SMAD/FHA domainSuperfamily: SMAD/FHA domainFamily: FHA domain26 c3fm8A_
not modelled
53.9
14
PDB header: transport protein/hydrolase activatorChain: A: PDB Molecule: kinesin-like protein kif13b;PDBTitle: crystal structure of full length centaurin alpha-1 bound with the fha2 domain of kif13b (capri target)
27 c3kf6B_
not modelled
47.4
20
PDB header: structural proteinChain: B: PDB Molecule: protein ten1;PDBTitle: crystal structure of s. pombe stn1-ten1 complex
28 c2eh0A_
not modelled
47.2
12
PDB header: transport proteinChain: A: PDB Molecule: kinesin-like protein kif1b;PDBTitle: solution structure of the fha domain from human kinesin-2 like protein kif1b
29 d1g6ga_
not modelled
46.7
15
Fold: SMAD/FHA domainSuperfamily: SMAD/FHA domainFamily: FHA domain30 d1dmza_
not modelled
43.4
17
Fold: SMAD/FHA domainSuperfamily: SMAD/FHA domainFamily: FHA domain31 d1kf6b2
not modelled
41.4
6
Fold: beta-Grasp (ubiquitin-like)Superfamily: 2Fe-2S ferredoxin-likeFamily: 2Fe-2S ferredoxin domains from multidomain proteins32 d1yjma1
not modelled
40.1
9
Fold: SMAD/FHA domainSuperfamily: SMAD/FHA domainFamily: FHA domain33 c3hx1B_
not modelled
38.4
10
PDB header: structural genomics, unknown functionChain: B: PDB Molecule: slr1951 protein;PDBTitle: crystal structure of the slr1951 protein from synechocystis sp.2 northeast structural genomics consortium target sgr167a
34 c1zeqX_
not modelled
38.0
15
PDB header: metal binding proteinChain: X: PDB Molecule: cation efflux system protein cusf;PDBTitle: 1.5 a structure of apo-cusf residues 6-88 from escherichia2 coli
35 c2o55A_
not modelled
37.6
22
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: putative glycerophosphodiester phosphodiesterase;PDBTitle: crystal structure of a putative glycerophosphodiester2 phosphodiesterase from galdieria sulphuraria
36 d1dm9a_
not modelled
36.4
11
Fold: Alpha-L RNA-binding motifSuperfamily: Alpha-L RNA-binding motifFamily: Heat shock protein 15 kD37 c1dm9A_
not modelled
36.4
11
PDB header: structural genomicsChain: A: PDB Molecule: hypothetical 15.5 kd protein in mrca-pckaPDBTitle: heat shock protein 15 kd
38 d2cu3a1
not modelled
34.5
13
Fold: beta-Grasp (ubiquitin-like)Superfamily: MoaD/ThiSFamily: ThiS39 c2k6pA_
not modelled
34.2
18
PDB header: unknown functionChain: A: PDB Molecule: uncharacterized protein hp_1423;PDBTitle: solution structure of hypothetical protein, hp1423
40 c2kklA_
not modelled
34.0
7
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: uncharacterized protein mb1858;PDBTitle: solution nmr structure of fha domain of mb1858 from2 mycobacterium bovis. northeast structural genomics3 consortium target mbr243c (24-155).
41 c3ks6A_
not modelled
30.9
18
PDB header: hydrolaseChain: A: PDB Molecule: glycerophosphoryl diester phosphodiesterase;PDBTitle: crystal structure of putative glycerophosphoryl diester2 phosphodiesterase (17743486) from agrobacterium tumefaciens3 str. c58 (dupont) at 1.80 a resolution
42 d1nekb2
not modelled
29.8
6
Fold: beta-Grasp (ubiquitin-like)Superfamily: 2Fe-2S ferredoxin-likeFamily: 2Fe-2S ferredoxin domains from multidomain proteins43 c2l55A_
not modelled
29.8
14
PDB header: metal binding proteinChain: A: PDB Molecule: silb,silver efflux protein, mfp component of the threePDBTitle: solution structure of the c-terminal domain of silb from cupriavidus2 metallidurans
44 c3gqsB_
not modelled
29.6
12
PDB header: structural genomics, unknown functionChain: B: PDB Molecule: adenylate cyclase-like protein;PDBTitle: crystal structure of the fha domain of ct664 protein from chlamydia2 trachomatis
45 d1snoa_
not modelled
28.2
19
Fold: OB-foldSuperfamily: Staphylococcal nucleaseFamily: Staphylococcal nuclease46 d2affa1
not modelled
27.9
9
Fold: SMAD/FHA domainSuperfamily: SMAD/FHA domainFamily: FHA domain47 d2ff4a3
not modelled
27.6
5
Fold: SMAD/FHA domainSuperfamily: SMAD/FHA domainFamily: FHA domain48 d1w96c1
not modelled
27.5
13
Fold: Barrel-sandwich hybridSuperfamily: Rudiment single hybrid motifFamily: BC C-terminal domain-like49 d1rkna_
not modelled
26.5
17
Fold: OB-foldSuperfamily: Staphylococcal nucleaseFamily: Staphylococcal nuclease50 d1mzka_
not modelled
26.4
9
Fold: SMAD/FHA domainSuperfamily: SMAD/FHA domainFamily: FHA domain51 c2jqlA_
not modelled
25.6
13
PDB header: cell cycleChain: A: PDB Molecule: dna damage response protein kinase dun1;PDBTitle: nmr structure of the yeast dun1 fha domain in complex with2 a doubly phosphorylated (pt) peptide derived from rad533 scd1
52 c3uotB_
not modelled
25.6
3
PDB header: cell cycleChain: B: PDB Molecule: mediator of dna damage checkpoint protein 1;PDBTitle: crystal structure of mdc1 fha domain in complex with a phosphorylated2 peptide from the mdc1 n-terminus
53 c3rlgA_
not modelled
25.3
11
PDB header: hydrolaseChain: A: PDB Molecule: sphingomyelin phosphodiesterase d lisictox-alphaia1a;PDBTitle: crystal structure of loxosceles intermedia phospholipase d isoform 12 h12a mutant
54 d1zud21
not modelled
24.7
14
Fold: beta-Grasp (ubiquitin-like)Superfamily: MoaD/ThiSFamily: ThiS55 c2zplA_
not modelled
24.3
26
PDB header: hydrolaseChain: A: PDB Molecule: regulator of sigma e protease;PDBTitle: crystal structure analysis of pdz domain a
56 d2oyza1
not modelled
24.2
12
Fold: Double-stranded beta-helixSuperfamily: RmlC-like cupinsFamily: VPA0057-like57 c1r21A_
not modelled
23.9
9
PDB header: cell cycleChain: A: PDB Molecule: antigen ki-67;PDBTitle: solution structure of human ki67 fha domain
58 d1lgpa_
not modelled
22.9
10
Fold: SMAD/FHA domainSuperfamily: SMAD/FHA domainFamily: FHA domain59 c3rggD_
not modelled
22.9
28
PDB header: lyaseChain: D: PDB Molecule: phosphoribosylaminoimidazole carboxylase, pure protein;PDBTitle: crystal structure of treponema denticola pure bound to air
60 d2bs2b2
not modelled
22.6
13
Fold: beta-Grasp (ubiquitin-like)Superfamily: 2Fe-2S ferredoxin-likeFamily: 2Fe-2S ferredoxin domains from multidomain proteins61 c2g1eA_
not modelled
22.5
15
PDB header: transferaseChain: A: PDB Molecule: hypothetical protein ta0895;PDBTitle: solution structure of ta0895
62 c2kl0A_
not modelled
22.4
24
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: putative thiamin biosynthesis this;PDBTitle: solution nmr structure of rhodopseudomonas palustris rpa3574,2 northeast structural genomics consortium (nesg) target rpr325
63 c2nnzA_
not modelled
21.5
13
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: hypothetical protein;PDBTitle: solution structure of the hypothetical protein af2241 from2 archaeoglobus fulgidus
64 c3eo6B_
not modelled
21.2
17
PDB header: structural genomics, unknown functionChain: B: PDB Molecule: protein of unknown function (duf1255);PDBTitle: crystal structure of protein of unknown function (duf1255)2 (afe_2634) from acidithiobacillus ferrooxidans ncib8455 at3 0.97 a resolution
65 d1wlna1
not modelled
21.0
12
Fold: SMAD/FHA domainSuperfamily: SMAD/FHA domainFamily: FHA domain66 d1k92a1
not modelled
21.0
33
Fold: Adenine nucleotide alpha hydrolase-likeSuperfamily: Adenine nucleotide alpha hydrolases-likeFamily: N-type ATP pyrophosphatases67 c3cwiA_
not modelled
20.5
13
PDB header: biosynthetic proteinChain: A: PDB Molecule: thiamine-biosynthesis protein this;PDBTitle: crystal structure of thiamine biosynthesis protein (this)2 from geobacter metallireducens. northeast structural3 genomics consortium target gmr137
68 c3rleA_
not modelled
20.5
33
PDB header: membrane proteinChain: A: PDB Molecule: golgi reassembly-stacking protein 2;PDBTitle: crystal structure of grasp55 grasp domain (residues 7-208)
69 d2z1ca1
not modelled
18.9
9
Fold: OB-foldSuperfamily: HupF/HypC-likeFamily: HupF/HypC-like70 d1v10a2
not modelled
17.7
13
Fold: Cupredoxin-likeSuperfamily: CupredoxinsFamily: Multidomain cupredoxins71 c3i10A_
not modelled
16.9
31
PDB header: hydrolaseChain: A: PDB Molecule: putative glycerophosphoryl diester phosphodiesterase;PDBTitle: crystal structure of putative glycerophosphoryl diester2 phosphodiesterase (np_812074.1) from bacteroides thetaiotaomicron3 vpi-5482 at 1.35 a resolution
72 c2jrtA_
not modelled
16.4
23
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: uncharacterized protein;PDBTitle: nmr solution structure of the protein coded by gene2 rhos4_12090 of rhodobacter sphaeroides. northeast3 structural genomics target rhr5
73 d2soba_
not modelled
15.9
16
Fold: OB-foldSuperfamily: Staphylococcal nucleaseFamily: Staphylococcal nuclease74 c3hvzB_
not modelled
15.0
23
PDB header: structural genomics, unknown functionChain: B: PDB Molecule: uncharacterized protein;PDBTitle: crystal structure of the tgs domain of the clolep_03100 protein from2 clostridium leptum, northeast structural genomics consortium target3 qlr13a
75 c3ju0A_
not modelled
14.8
38
PDB header: dna binding proteinChain: A: PDB Molecule: phage integrase;PDBTitle: structure of the arm-type binding domain of hai7 integrase
76 d1u1ha1
not modelled
14.6
44
Fold: TIM beta/alpha-barrelSuperfamily: UROD/MetE-likeFamily: Cobalamin-independent methionine synthase77 c3r6oA_
not modelled
14.3
30
PDB header: isomeraseChain: A: PDB Molecule: 2-hydroxyhepta-2,4-diene-1, 7-dioateisomerase;PDBTitle: crystal structure of a probable 2-hydroxyhepta-2,4-diene-1, 7-2 dioateisomerase from mycobacterium abscessus
78 d1zcca1
not modelled
14.1
23
Fold: TIM beta/alpha-barrelSuperfamily: PLC-like phosphodiesterasesFamily: Glycerophosphoryl diester phosphodiesterase79 c2aanA_
not modelled
13.9
17
PDB header: electron transportChain: A: PDB Molecule: auracyanin a;PDBTitle: auracyanin a: a "blue" copper protein from the green thermophilic2 photosynthetic bacterium,chloroflexus aurantiacus
80 d2pyta1
not modelled
13.7
13
Fold: Double-stranded beta-helixSuperfamily: RmlC-like cupinsFamily: EutQ-like81 d1tygb_
not modelled
13.7
3
Fold: beta-Grasp (ubiquitin-like)Superfamily: MoaD/ThiSFamily: ThiS82 d2bh1x1
not modelled
13.5
13
Fold: Alpha-lytic protease prodomain-likeSuperfamily: EspE N-terminal domain-likeFamily: GSPII protein E N-terminal domain-like83 c2bh1Y_
not modelled
13.5
13
PDB header: transport proteinChain: Y: PDB Molecule: general secretion pathway protein e,;PDBTitle: x-ray structure of the general secretion pathway complex of2 the n-terminal domain of epse and the cytosolic domain of3 epsl of vibrio cholerae
84 c3rlhA_
not modelled
13.4
13
PDB header: hydrolaseChain: A: PDB Molecule: sphingomyelin phosphodiesterase d lisictox-alphaia1a;PDBTitle: crystal structure of a class ii phospholipase d from loxosceles2 intermedia venom
85 d1tkea1
not modelled
13.3
13
Fold: beta-Grasp (ubiquitin-like)Superfamily: TGS-likeFamily: TGS domain86 d1nyra2
not modelled
13.3
9
Fold: beta-Grasp (ubiquitin-like)Superfamily: TGS-likeFamily: TGS domain87 c2vgmA_
not modelled
13.2
17
PDB header: cell cycleChain: A: PDB Molecule: dom34;PDBTitle: structure of yeast dom34 : a protein related to translation2 termination factor erf1 and involved in no-go decay.
88 c1kh2D_
not modelled
13.0
35
PDB header: ligaseChain: D: PDB Molecule: argininosuccinate synthetase;PDBTitle: crystal structure of thermus thermophilus hb82 argininosuccinate synthetase in complex with atp
89 c1tygG_
not modelled
12.8
0
PDB header: biosynthetic proteinChain: G: PDB Molecule: yjbs;PDBTitle: structure of the thiazole synthase/this complex
90 c3rpfB_
not modelled
12.8
13
PDB header: transferaseChain: B: PDB Molecule: molybdopterin synthase catalytic subunit;PDBTitle: protein-protein complex of subunit 1 and 2 of molybdopterin-converting2 factor from helicobacter pylori 26695
91 d1j20a1
not modelled
12.5
35
Fold: Adenine nucleotide alpha hydrolase-likeSuperfamily: Adenine nucleotide alpha hydrolases-likeFamily: N-type ATP pyrophosphatases92 c3hqxA_
not modelled
12.4
16
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: upf0345 protein aciad0356;PDBTitle: crystal structure of protein of unknown function (duf1255,pf06865)2 from acinetobacter sp. adp1
93 d2oa4a1
not modelled
12.4
25
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: TrpR-likeFamily: SPO1678-like94 d1vl2a1
not modelled
12.0
31
Fold: Adenine nucleotide alpha hydrolase-likeSuperfamily: Adenine nucleotide alpha hydrolases-likeFamily: N-type ATP pyrophosphatases95 c2zpmA_
not modelled
12.0
18
PDB header: hydrolaseChain: A: PDB Molecule: regulator of sigma e protease;PDBTitle: crystal structure analysis of pdz domain b
96 c2x4iA_
not modelled
11.6
6
PDB header: unknown functionChain: A: PDB Molecule: uncharacterized protein 114;PDBTitle: orf 114a from sulfolobus islandicus rudivirus 1
97 c3r7kB_
not modelled
11.6
13
PDB header: oxidoreductaseChain: B: PDB Molecule: probable acyl coa dehydrogenase;PDBTitle: crystal structure of a probable acyl coa dehydrogenase from2 mycobacterium abscessus atcc 19977 / dsm 44196
98 d1tt2a_
not modelled
11.5
19
Fold: OB-foldSuperfamily: Staphylococcal nucleaseFamily: Staphylococcal nuclease99 c3no3A_
not modelled
11.5
21
PDB header: hydrolaseChain: A: PDB Molecule: glycerophosphodiester phosphodiesterase;PDBTitle: crystal structure of a glycerophosphodiester phosphodiesterase2 (bdi_0402) from parabacteroides distasonis atcc 8503 at 1.89 a3 resolution