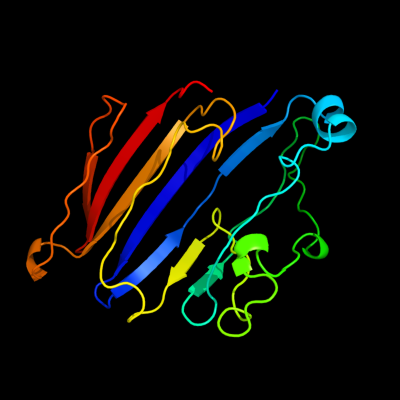
1 c3kd4A_
87.5
9
PDB header: hydrolaseChain: A: PDB Molecule: putative protease;PDBTitle: crystal structure of a putative protease (bdi_1141) from2 parabacteroides distasonis atcc 8503 at 2.00 a resolution
2 d3b7sa2
72.1
9
Fold: Leukotriene A4 hydrolase N-terminal domainSuperfamily: Leukotriene A4 hydrolase N-terminal domainFamily: Leukotriene A4 hydrolase N-terminal domain3 c3ebhA_
71.9
5
PDB header: hydrolase inhibitorChain: A: PDB Molecule: m1 family aminopeptidase;PDBTitle: structure of the m1 alanylaminopeptidase from malaria complexed with2 bestatin
4 c3b7uX_
61.6
10
PDB header: hydrolaseChain: X: PDB Molecule: leukotriene a-4 hydrolase;PDBTitle: leukotriene a4 hydrolase complexed with kelatorphan
5 c2gtqA_
52.9
12
PDB header: hydrolaseChain: A: PDB Molecule: aminopeptidase n;PDBTitle: crystal structure of aminopeptidase n from human pathogen neisseria2 meningitidis
6 c3ciaA_
51.5
9
PDB header: hydrolaseChain: A: PDB Molecule: cold-active aminopeptidase;PDBTitle: crystal structure of cold-aminopeptidase from colwellia2 psychrerythraea
7 c2xpyA_
35.6
10
PDB header: hydrolaseChain: A: PDB Molecule: leukotriene a-4 hydrolase;PDBTitle: structure of native leukotriene a4 hydrolase from saccharomyces2 cerevisiae
8 d2etha1
33.5
13
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: MarR-like transcriptional regulators9 d1tkna_
28.9
12
Fold: STAT-likeSuperfamily: CAPPD, an extracellular domain of amyloid beta A4 proteinFamily: CAPPD, an extracellular domain of amyloid beta A4 protein10 d1lj9a_
26.4
16
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: MarR-like transcriptional regulators11 d2fbha1
22.9
10
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: MarR-like transcriptional regulators12 c3hd7A_
21.8
8
PDB header: exocytosisChain: A: PDB Molecule: vesicle-associated membrane protein 2;PDBTitle: helical extension of the neuronal snare complex into the membrane,2 spacegroup c 1 2 1
13 c3se6A_
19.7
12
PDB header: hydrolaseChain: A: PDB Molecule: endoplasmic reticulum aminopeptidase 2;PDBTitle: crystal structure of the human endoplasmic reticulum aminopeptidase 2
14 c2i7uA_
19.5
100
PDB header: de novo protein/ligand binding proteinChain: A: PDB Molecule: four-alpha-helix bundle;PDBTitle: structural and dynamical analysis of a four-alpha-helix2 bundle with designed anesthetic binding pockets
15 c3nrvC_
18.3
13
PDB header: transcription regulatorChain: C: PDB Molecule: putative transcriptional regulator (marr/emrr family);PDBTitle: crystal structure of marr/emrr family transcriptional regulator from2 acinetobacter sp. adp1
16 d2a61a1
17.9
9
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: MarR-like transcriptional regulators17 c2rngA_
16.8
13
PDB header: antimicrobial proteinChain: A: PDB Molecule: big defensin;PDBTitle: solution structure of big defensin
18 c3g3zA_
15.3
12
PDB header: transcriptionChain: A: PDB Molecule: transcriptional regulator, marr family;PDBTitle: the structure of nmb1585, a marr family regulator from neisseria2 meningitidis
19 c2rh0B_
14.9
11
PDB header: nuclear proteinChain: B: PDB Molecule: nudc domain-containing protein 2;PDBTitle: crystal structure of nudc domain-containing protein 22 (13542905) from mus musculus at 1.95 a resolution
20 d1xp8a2
12.0
16
Fold: Anti-LPS factor/recA domainSuperfamily: RecA protein, C-terminal domainFamily: RecA protein, C-terminal domain21 d1s3ja_
not modelled
11.1
7
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: MarR-like transcriptional regulators22 c3bj6B_
not modelled
11.0
11
PDB header: transcription regulatorChain: B: PDB Molecule: transcriptional regulator, marr family;PDBTitle: crystal structure of marr family transcription regulator sp03579
23 d1rw6a_
not modelled
10.9
12
Fold: STAT-likeSuperfamily: CAPPD, an extracellular domain of amyloid beta A4 proteinFamily: CAPPD, an extracellular domain of amyloid beta A4 protein24 c3k66A_
not modelled
10.6
19
PDB header: cell adhesionChain: A: PDB Molecule: beta-amyloid-like protein;PDBTitle: x-ray crystal structure of the e2 domain of c. elegans apl-1
25 c3bogC_
not modelled
10.3
87
PDB header: antifreeze proteinChain: C: PDB Molecule: 6.5 kda glycine-rich antifreeze protein;PDBTitle: snow flea antifreeze protein quasi-racemate
26 c3bogD_
not modelled
10.3
87
PDB header: antifreeze proteinChain: D: PDB Molecule: 6.5 kda glycine-rich antifreeze protein;PDBTitle: snow flea antifreeze protein quasi-racemate
27 c1z5hB_
not modelled
10.2
12
PDB header: hydrolaseChain: B: PDB Molecule: tricorn protease interacting factor f3;PDBTitle: crystal structures of the tricorn interacting factor f32 from thermoplasma acidophilum
28 c3bjaA_
not modelled
9.7
11
PDB header: transcription regulatorChain: A: PDB Molecule: transcriptional regulator, marr family, putative;PDBTitle: crystal structure of putative marr-like transcription regulator2 (np_978771.1) from bacillus cereus atcc 10987 at 2.38 a resolution
29 c3ju0A_
not modelled
9.6
18
PDB header: dna binding proteinChain: A: PDB Molecule: phage integrase;PDBTitle: structure of the arm-type binding domain of hai7 integrase
30 c2zouB_
not modelled
9.2
21
PDB header: cell adhesionChain: B: PDB Molecule: spondin-1;PDBTitle: crystal struture of human f-spondin reeler domain (fragment 2)
31 c2vn2B_
not modelled
9.0
19
PDB header: replicationChain: B: PDB Molecule: chromosome replication initiation protein;PDBTitle: crystal structure of the n-terminal domain of dnad protein2 from geobacillus kaustophilus hta426
32 d2v9va2
not modelled
7.9
15
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: C-terminal fragment of elongation factor SelB33 c3cwcB_
not modelled
7.8
35
PDB header: transferaseChain: B: PDB Molecule: putative glycerate kinase 2;PDBTitle: crystal structure of putative glycerate kinase 2 from salmonella2 typhimurium lt2
34 c3jtzA_
not modelled
7.4
21
PDB header: dna binding proteinChain: A: PDB Molecule: integrase;PDBTitle: structure of the arm-type binding domain of hpi integrase
35 c1cn3F_
not modelled
7.1
88
PDB header: viral proteinChain: F: PDB Molecule: fragment of coat protein vp2;PDBTitle: interaction of polyomavirus internal protein vp2 with major2 capsid protein vp1 and implications for participation of3 vp2 in viral entry
36 c2fl8N_
not modelled
6.6
10
PDB header: virus/viral proteinChain: N: PDB Molecule: baseplate structural protein gp10;PDBTitle: fitting of the gp10 trimer structure into the cryoem map of the2 bacteriophage t4 baseplate in the hexagonal conformation.
37 c2pqnB_
not modelled
6.6
27
PDB header: apoptosisChain: B: PDB Molecule: mitochondrial division protein 1;PDBTitle: crystal structure of yeast fis1 complexed with a fragment of yeast2 mdv1
38 c3c5pF_
not modelled
6.6
16
PDB header: structural genomics, unknown functionChain: F: PDB Molecule: protein bas0735 of unknown function;PDBTitle: crystal structure of bas0735, a protein of unknown function from2 bacillus anthracis str. sterne
39 c3cooB_
not modelled
6.4
21
PDB header: cell adhesionChain: B: PDB Molecule: spondin-1;PDBTitle: the crystal structure of reelin-n domain of f-spondin
40 c2oq0D_
not modelled
6.3
16
PDB header: protein bindingChain: D: PDB Molecule: gamma-interferon-inducible protein ifi-16;PDBTitle: crystal structure of the first hin-200 domain of interferon-inducible2 protein 16
41 c3pa8A_
not modelled
6.2
9
PDB header: toxin/peptide inhibitorChain: A: PDB Molecule: toxin b;PDBTitle: structure of the c. difficile tcdb cysteine protease domain in complex2 with a peptide inhibitor
42 c2l3nA_
not modelled
6.2
53
PDB header: dna binding proteinChain: A: PDB Molecule: dna-binding protein rap1, telomere length regulator taz1;PDBTitle: solution structure of rap1-taz1 fusion protein
43 c2rddB_
not modelled
6.0
7
PDB header: membrane protein/transport proteinChain: B: PDB Molecule: upf0092 membrane protein yajc;PDBTitle: x-ray crystal structure of acrb in complex with a novel2 transmembrane helix.
44 c2khuA_
not modelled
6.0
25
PDB header: transferase/protein bindingChain: A: PDB Molecule: immunoglobulin g-binding protein g, dnaPDBTitle: solution structure of the ubiquitin-binding motif of human2 polymerase iota
45 c2l0gA_
not modelled
5.9
25
PDB header: protein bindingChain: A: PDB Molecule: dna polymerase iota;PDBTitle: solution nmr structure of ubiquitin-binding motif (ubm2) of human2 polymerase iota
46 c3pmrB_
not modelled
5.9
27
PDB header: cell adhesionChain: B: PDB Molecule: amyloid-like protein 1;PDBTitle: crystal structure of e2 domain of human amyloid precursor-like protein2 1
47 d2hyma1
not modelled
5.8
9
Fold: Immunoglobulin-like beta-sandwichSuperfamily: Fibronectin type IIIFamily: Fibronectin type III48 c2nnnB_
not modelled
5.8
12
PDB header: transcriptionChain: B: PDB Molecule: probable transcriptional regulator;PDBTitle: crystal structure of probable transcriptional regulator from2 pseudomonas aeruginosa
49 d1wfia_
not modelled
5.6
7
Fold: HSP20-like chaperonesSuperfamily: HSP20-like chaperonesFamily: Nuclear movement domain50 c3n39D_
not modelled
5.5
14
PDB header: oxidoreductaseChain: D: PDB Molecule: protein nrdi;PDBTitle: ribonucleotide reductase dimanganese(ii)-nrdf from escherichia coli in2 complex with nrdi
51 c2x8kB_
not modelled
5.3
12
PDB header: viral proteinChain: B: PDB Molecule: hypothetical protein 19.1;PDBTitle: crystal structure of spp1 dit (gp 19.1) protein, a paradigm2 of hub adsorption apparatus in gram-positive infecting3 phages.
52 c2o30A_
not modelled
5.3
9
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: nuclear movement protein;PDBTitle: nuclear movement protein from e. cuniculi gb-m1
53 c2kncA_
not modelled
5.3
18
PDB header: cell adhesionChain: A: PDB Molecule: integrin alpha-iib;PDBTitle: platelet integrin alfaiib-beta3 transmembrane-cytoplasmic2 heterocomplex
54 c2jo1A_
not modelled
5.3
23
PDB header: hydrolase regulatorChain: A: PDB Molecule: phospholemman;PDBTitle: structure of the na,k-atpase regulatory protein fxyd1 in2 micelles
55 c1kn7A_
not modelled
5.2
30
PDB header: membrane proteinChain: A: PDB Molecule: voltage-gated potassium channel protein kv1.4;PDBTitle: solution structure of the tandem inactivation domain2 (residues 1-75) of potassium channel rck4 (kv1.4)
56 d1ubea2
not modelled
5.2
16
Fold: Anti-LPS factor/recA domainSuperfamily: RecA protein, C-terminal domainFamily: RecA protein, C-terminal domain57 d1mo6a2
not modelled
5.1
16
Fold: Anti-LPS factor/recA domainSuperfamily: RecA protein, C-terminal domainFamily: RecA protein, C-terminal domain58 c2qwwB_
not modelled
5.0
13
PDB header: transcriptionChain: B: PDB Molecule: transcriptional regulator, marr family;PDBTitle: crystal structure of multiple antibiotic-resistance repressor (marr)2 (yp_013417.1) from listeria monocytogenes 4b f2365 at 2.07 a3 resolution