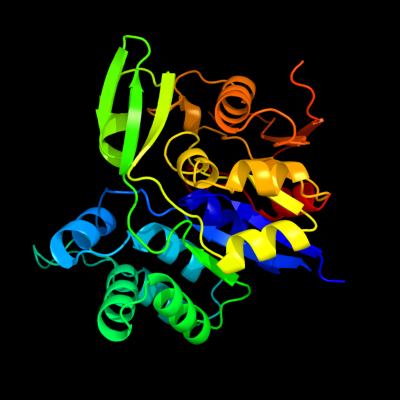
| 1 | d2hzba1
|
|
 |
100.0 |
30 |
Fold:CofD-like
Superfamily:CofD-like
Family:CofD-like |
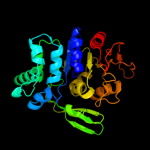
| 2 | c2ppvA_
|
|
 |
100.0 |
34 |
PDB header:transferase
Chain: A: PDB Molecule:uncharacterized protein;
PDBTitle: crystal structure of a protein belonging to the upf0052 (se_0549) from2 staphylococcus epidermidis atcc 12228 at 2.00 a resolution
|
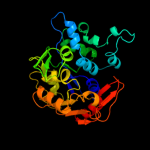
| 3 | c2q7xA_
|
|
 |
100.0 |
29 |
PDB header:transferase
Chain: A: PDB Molecule:upf0052 protein sp_1565;
PDBTitle: crystal structure of a putative phospho transferase (sp_1565) from2 streptococcus pneumoniae tigr4 at 2.00 a resolution
|
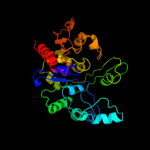
| 4 | d2ffea1
|
|
 |
100.0 |
16 |
Fold:CofD-like
Superfamily:CofD-like
Family:CofD-like |
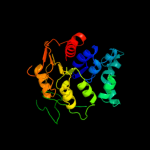
| 5 | c2p0yA_
|
|
 |
100.0 |
29 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:hypothetical protein lp_0780;
PDBTitle: crystal structure of q88yi3_lacpl from lactobacillus2 plantarum. northeast structural genomics consortium target3 lpr6
|
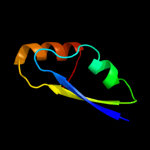
| 6 | c3dzcA_
|
|
 |
88.7 |
8 |
PDB header:isomerase
Chain: A: PDB Molecule:udp-n-acetylglucosamine 2-epimerase;
PDBTitle: 2.35 angstrom resolution structure of wecb (vc0917), a udp-n-2 acetylglucosamine 2-epimerase from vibrio cholerae.
|
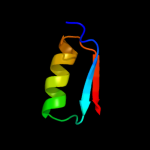
| 7 | d1p3da1
|
|
 |
86.8 |
21 |
Fold:MurCD N-terminal domain
Superfamily:MurCD N-terminal domain
Family:MurCD N-terminal domain |
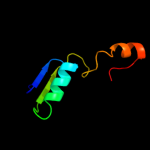
| 8 | c3kljA_
|
|
 |
86.7 |
13 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:nad(fad)-dependent dehydrogenase, nirb-family (n-terminal
PDBTitle: crystal structure of nadh:rubredoxin oxidoreductase from clostridium2 acetobutylicum
|
| 9 | c3kpgA_
|
|
 |
86.5 |
33 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:sulfide-quinone reductase, putative;
PDBTitle: crystal structure of sulfide:quinone oxidoreductase from2 acidithiobacillus ferrooxidans in complex with decylubiquinone
|
| 10 | d2afba1
|
|
 |
84.9 |
24 |
Fold:Ribokinase-like
Superfamily:Ribokinase-like
Family:Ribokinase-like |
| 11 | c1yqzA_
|
|
 |
84.4 |
15 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:coenzyme a disulfide reductase;
PDBTitle: structure of coenzyme a-disulfide reductase from2 staphylococcus aureus refined at 1.54 angstrom resolution
|
| 12 | d2blna2
|
|
 |
82.9 |
23 |
Fold:Formyltransferase
Superfamily:Formyltransferase
Family:Formyltransferase |
| 13 | d1fcda1
|
|
 |
82.2 |
19 |
Fold:FAD/NAD(P)-binding domain
Superfamily:FAD/NAD(P)-binding domain
Family:FAD/NAD-linked reductases, N-terminal and central domains |
| 14 | c1yrwA_
|
|
 |
82.0 |
23 |
PDB header:transferase
Chain: A: PDB Molecule:protein arna;
PDBTitle: crystal structure of e.coli arna transformylase domain
|
| 15 | c1kyqC_
|
|
 |
81.5 |
24 |
PDB header:oxidoreductase, lyase
Chain: C: PDB Molecule:siroheme biosynthesis protein met8;
PDBTitle: met8p: a bifunctional nad-dependent dehydrogenase and2 ferrochelatase involved in siroheme synthesis.
|
| 16 | c1fcdB_
|
|
 |
81.2 |
19 |
PDB header:electron transport(flavocytochrome)
Chain: B: PDB Molecule:flavocytochrome c sulfide dehydrogenase (flavin-
PDBTitle: the structure of flavocytochrome c sulfide dehydrogenase2 from a purple phototrophic bacterium chromatium vinosum at3 2.5 angstroms resolution
|
| 17 | c3dfzB_
|
|
 |
81.1 |
22 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:precorrin-2 dehydrogenase;
PDBTitle: sirc, precorrin-2 dehydrogenase
|
| 18 | c2e4gB_
|
|
 |
80.9 |
26 |
PDB header:biosynthetic protein, flavoprotein
Chain: B: PDB Molecule:tryptophan halogenase;
PDBTitle: rebh with bound l-trp
|
| 19 | d1fmta2
|
|
 |
80.9 |
21 |
Fold:Formyltransferase
Superfamily:Formyltransferase
Family:Formyltransferase |
| 20 | d1f0ka_
|
|
 |
80.7 |
24 |
Fold:UDP-Glycosyltransferase/glycogen phosphorylase
Superfamily:UDP-Glycosyltransferase/glycogen phosphorylase
Family:Peptidoglycan biosynthesis glycosyltransferase MurG |
| 21 | c3n8kG_ |
|
not modelled |
79.2 |
25 |
PDB header:lyase
Chain: G: PDB Molecule:3-dehydroquinate dehydratase;
PDBTitle: type ii dehydroquinase from mycobacterium tuberculosis complexed with2 citrazinic acid
|
| 22 | d1sb8a_ |
|
not modelled |
78.9 |
19 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Tyrosine-dependent oxidoreductases |
| 23 | c3ia7A_ |
|
not modelled |
77.1 |
18 |
PDB header:transferase
Chain: A: PDB Molecule:calg4;
PDBTitle: crystal structure of calg4, the calicheamicin glycosyltransferase
|
| 24 | c2weuD_ |
|
not modelled |
77.0 |
27 |
PDB header:antifungal protein
Chain: D: PDB Molecule:tryptophan 5-halogenase;
PDBTitle: crystal structure of tryptophan 5-halogenase (pyrh) complex2 with substrate tryptophan
|
| 25 | d1kyqa1 |
|
not modelled |
76.9 |
24 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Siroheme synthase N-terminal domain-like |
| 26 | c3allA_ |
|
not modelled |
76.6 |
19 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:2-methyl-3-hydroxypyridine-5-carboxylic acid oxygenase;
PDBTitle: crystal structure of 2-methyl-3-hydroxypyridine-5-carboxylic acid2 oxygenase, mutant y270a
|
| 27 | d1pj5a2 |
|
not modelled |
75.7 |
11 |
Fold:FAD/NAD(P)-binding domain
Superfamily:FAD/NAD(P)-binding domain
Family:FAD-linked reductases, N-terminal domain |
| 28 | c3kd9B_ |
|
not modelled |
75.4 |
28 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:coenzyme a disulfide reductase;
PDBTitle: crystal structure of pyridine nucleotide disulfide oxidoreductase from2 pyrococcus horikoshii
|
| 29 | c2vdcI_
|
|
 |
75.4 |
16 |
PDB header:oxidoreductase
Chain: I: PDB Molecule:glutamate synthase [nadph] small chain;
PDBTitle: the 9.5 a resolution structure of glutamate synthase from2 cryo-electron microscopy and its oligomerization behavior3 in solution: functional implications.
|
| 30 | d1q1ra2 |
|
not modelled |
75.4 |
25 |
Fold:FAD/NAD(P)-binding domain
Superfamily:FAD/NAD(P)-binding domain
Family:FAD/NAD-linked reductases, N-terminal and central domains |
| 31 | c1pjtB_ |
|
not modelled |
74.5 |
27 |
PDB header:transferase/oxidoreductase/lyase
Chain: B: PDB Molecule:siroheme synthase;
PDBTitle: the structure of the ser128ala point-mutant variant of cysg,2 the multifunctional3 methyltransferase/dehydrogenase/ferrochelatase for4 siroheme synthesis
|
| 32 | d1ndha2 |
|
not modelled |
74.1 |
20 |
Fold:Ferredoxin reductase-like, C-terminal NADP-linked domain
Superfamily:Ferredoxin reductase-like, C-terminal NADP-linked domain
Family:Reductases |
| 33 | c3ps9A_ |
|
not modelled |
74.1 |
23 |
PDB header:transferase
Chain: A: PDB Molecule:trna 5-methylaminomethyl-2-thiouridine biosynthesis
PDBTitle: crystal structure of mnmc from e. coli
|
| 34 | c1ps9A_ |
|
not modelled |
73.8 |
13 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:2,4-dienoyl-coa reductase;
PDBTitle: the crystal structure and reaction mechanism of e. coli 2,4-2 dienoyl coa reductase
|
| 35 | c2uygF_ |
|
not modelled |
73.6 |
26 |
PDB header:lyase
Chain: F: PDB Molecule:3-dehydroquinate dehydratase;
PDBTitle: crystallogaphic structure of the typeii 3-dehydroquinase2 from thermus thermophilus
|
| 36 | d1cjca2 |
|
not modelled |
73.2 |
17 |
Fold:Nucleotide-binding domain
Superfamily:Nucleotide-binding domain
Family:N-terminal domain of adrenodoxin reductase-like |
| 37 | c3icrA_ |
|
not modelled |
73.2 |
24 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:coenzyme a-disulfide reductase;
PDBTitle: crystal structure of oxidized bacillus anthracis coadr-rhd
|
| 38 | c3i3lA_ |
|
not modelled |
72.7 |
26 |
PDB header:hydrolase
Chain: A: PDB Molecule:alkylhalidase cmls;
PDBTitle: crystal structure of cmls, a flavin-dependent halogenase
|
| 39 | c3lxdA_ |
|
not modelled |
72.6 |
30 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:fad-dependent pyridine nucleotide-disulphide
PDBTitle: crystal structure of ferredoxin reductase arr from novosphingobium2 aromaticivorans
|
| 40 | d1vdca2 |
|
not modelled |
71.9 |
21 |
Fold:FAD/NAD(P)-binding domain
Superfamily:FAD/NAD(P)-binding domain
Family:FAD/NAD-linked reductases, N-terminal and central domains |
| 41 | d1b5qa1 |
|
not modelled |
71.7 |
28 |
Fold:FAD/NAD(P)-binding domain
Superfamily:FAD/NAD(P)-binding domain
Family:FAD-linked reductases, N-terminal domain |
| 42 | c1h83A_ |
|
not modelled |
71.1 |
28 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:polyamine oxidase;
PDBTitle: structure of polyamine oxidase in complex with2 1,8-diaminooctane
|
| 43 | d1h05a_ |
|
not modelled |
70.8 |
25 |
Fold:Flavodoxin-like
Superfamily:Type II 3-dehydroquinate dehydratase
Family:Type II 3-dehydroquinate dehydratase |
| 44 | c1c0iA_ |
|
not modelled |
70.5 |
24 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:d-amino acid oxidase;
PDBTitle: crystal structure of d-amino acid oxidase in complex with2 two anthranylate molecules
|
| 45 | d1trba2 |
|
not modelled |
70.3 |
24 |
Fold:FAD/NAD(P)-binding domain
Superfamily:FAD/NAD(P)-binding domain
Family:FAD/NAD-linked reductases, N-terminal and central domains |
| 46 | c1k97A_ |
|
not modelled |
70.1 |
19 |
PDB header:ligase
Chain: A: PDB Molecule:argininosuccinate synthase;
PDBTitle: crystal structure of e. coli argininosuccinate synthetase in complex2 with aspartate and citrulline
|
| 47 | d2dw4a2 |
|
not modelled |
70.1 |
23 |
Fold:FAD/NAD(P)-binding domain
Superfamily:FAD/NAD(P)-binding domain
Family:FAD-linked reductases, N-terminal domain |
| 48 | d1fl2a2 |
|
not modelled |
69.9 |
15 |
Fold:FAD/NAD(P)-binding domain
Superfamily:FAD/NAD(P)-binding domain
Family:FAD/NAD-linked reductases, N-terminal and central domains |
| 49 | d1fdra2 |
|
not modelled |
69.3 |
20 |
Fold:Ferredoxin reductase-like, C-terminal NADP-linked domain
Superfamily:Ferredoxin reductase-like, C-terminal NADP-linked domain
Family:Reductases |
| 50 | c3qj4A_ |
|
not modelled |
69.3 |
19 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:renalase;
PDBTitle: crystal structure of human renalase (isoform 1)
|
| 51 | c3ntaA_ |
|
not modelled |
69.1 |
16 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:fad-dependent pyridine nucleotide-disulphide
PDBTitle: structure of the shewanella loihica pv-4 nadh-dependent persulfide2 reductase
|
| 52 | d1uqra_ |
|
not modelled |
69.1 |
39 |
Fold:Flavodoxin-like
Superfamily:Type II 3-dehydroquinate dehydratase
Family:Type II 3-dehydroquinate dehydratase |
| 53 | d1c0pa1 |
|
not modelled |
68.8 |
24 |
Fold:Nucleotide-binding domain
Superfamily:Nucleotide-binding domain
Family:D-aminoacid oxidase, N-terminal domain |
| 54 | c3g5rA_ |
|
not modelled |
68.8 |
23 |
PDB header:transferase
Chain: A: PDB Molecule:methylenetetrahydrofolate--trna-(uracil-5-)-
PDBTitle: crystal structure of thermus thermophilus trmfo in complex with2 tetrahydrofolate
|
| 55 | c2v3aA_ |
|
not modelled |
68.8 |
13 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:rubredoxin reductase;
PDBTitle: crystal structure of rubredoxin reductase from pseudomonas2 aeruginosa.
|
| 56 | c3hyxC_ |
|
not modelled |
68.1 |
33 |
PDB header:oxidoreductase
Chain: C: PDB Molecule:sulfide-quinone reductase;
PDBTitle: 3-d x-ray structure of the sulfide:quinone oxidoreductase from aquifex2 aeolicus in complex with aurachin c
|
| 57 | d1gqoa_ |
|
not modelled |
67.7 |
25 |
Fold:Flavodoxin-like
Superfamily:Type II 3-dehydroquinate dehydratase
Family:Type II 3-dehydroquinate dehydratase |
| 58 | c2cmgA_ |
|
not modelled |
67.3 |
15 |
PDB header:transferase
Chain: A: PDB Molecule:spermidine synthase;
PDBTitle: crystal structure of spermidine synthase from helicobacter2 pylori
|
| 59 | d1d7ya2 |
|
not modelled |
67.2 |
17 |
Fold:FAD/NAD(P)-binding domain
Superfamily:FAD/NAD(P)-binding domain
Family:FAD/NAD-linked reductases, N-terminal and central domains |
| 60 | d1ep3b2 |
|
not modelled |
66.9 |
21 |
Fold:Ferredoxin reductase-like, C-terminal NADP-linked domain
Superfamily:Ferredoxin reductase-like, C-terminal NADP-linked domain
Family:Dihydroorotate dehydrogenase B, PyrK subunit |
| 61 | d1k92a1 |
|
not modelled |
66.8 |
20 |
Fold:Adenine nucleotide alpha hydrolase-like
Superfamily:Adenine nucleotide alpha hydrolases-like
Family:N-type ATP pyrophosphatases |
| 62 | c3djeA_ |
|
not modelled |
66.4 |
15 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:fructosyl amine: oxygen oxidoreductase;
PDBTitle: crystal structure of the deglycating enzyme fructosamine2 oxidase from aspergillus fumigatus (amadoriase ii) in3 complex with fsa
|
| 63 | c2ardA_ |
|
not modelled |
66.3 |
29 |
PDB header:biosynthetic protein
Chain: A: PDB Molecule:tryptophan halogenase prna;
PDBTitle: the structure of tryptophan 7-halogenase (prna) suggests a mechanism2 for regioselective chlorination
|
| 64 | c3ab1B_ |
|
not modelled |
66.0 |
13 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:ferredoxin--nadp reductase;
PDBTitle: crystal structure of ferredoxin nadp+ oxidoreductase
|
| 65 | c3ihmB_ |
|
not modelled |
65.3 |
18 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:styrene monooxygenase a;
PDBTitle: structure of the oxygenase component of a pseudomonas styrene2 monooxygenase
|
| 66 | d1onfa2 |
|
not modelled |
65.3 |
12 |
Fold:FAD/NAD(P)-binding domain
Superfamily:FAD/NAD(P)-binding domain
Family:FAD/NAD-linked reductases, N-terminal and central domains |
| 67 | c3k5iB_ |
|
not modelled |
65.3 |
11 |
PDB header:lyase
Chain: B: PDB Molecule:phosphoribosyl-aminoimidazole carboxylase;
PDBTitle: crystal structure of n5-carboxyaminoimidazole synthase from2 aspergillus clavatus in complex with adp and 5-3 aminoimadazole ribonucleotide
|
| 68 | c3k30B_ |
|
not modelled |
65.1 |
20 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:histamine dehydrogenase;
PDBTitle: histamine dehydrogenase from nocardiodes simplex
|
| 69 | c2gr2A_ |
|
not modelled |
65.1 |
26 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:ferredoxin reductase;
PDBTitle: crystal structure of ferredoxin reductase, bpha4 (oxidized form)
|
| 70 | c3iwaA_ |
|
not modelled |
65.0 |
13 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:fad-dependent pyridine nucleotide-disulphide
PDBTitle: crystal structure of a fad-dependent pyridine nucleotide-disulphide2 oxidoreductase from desulfovibrio vulgaris
|
| 71 | c3lrxC_ |
|
not modelled |
64.7 |
16 |
PDB header:oxidoreductase
Chain: C: PDB Molecule:putative hydrogenase;
PDBTitle: crystal structure of the c-terminal domain (residues 78-226)2 of pf1911 hydrogenase from pyrococcus furiosus, northeast3 structural genomics consortium target pfr246a
|
| 72 | c1djnB_ |
|
not modelled |
64.5 |
20 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:trimethylamine dehydrogenase;
PDBTitle: structural and biochemical characterization of recombinant wild type2 trimethylamine dehydrogenase from methylophilus methylotrophus (sp.3 w3a1)
|
| 73 | c2b9yA_ |
|
not modelled |
64.4 |
18 |
PDB header:isomerase
Chain: A: PDB Molecule:putative aminooxidase;
PDBTitle: crystal structure of cla-producing fatty acid isomerase2 from p. acnes
|
| 74 | c3ef6A_ |
|
not modelled |
64.3 |
21 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:toluene 1,2-dioxygenase system ferredoxin--nad(+)
PDBTitle: crystal structure of toluene 2,3-dioxygenase reductase
|
| 75 | c1q1wA_ |
|
not modelled |
64.1 |
25 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:putidaredoxin reductase;
PDBTitle: crystal structure of putidaredoxin reductase from2 pseudomonas putida
|
| 76 | d1qx4a2 |
|
not modelled |
63.9 |
20 |
Fold:Ferredoxin reductase-like, C-terminal NADP-linked domain
Superfamily:Ferredoxin reductase-like, C-terminal NADP-linked domain
Family:Reductases |
| 77 | d1seza1 |
|
not modelled |
63.9 |
23 |
Fold:FAD/NAD(P)-binding domain
Superfamily:FAD/NAD(P)-binding domain
Family:FAD-linked reductases, N-terminal domain |
| 78 | c2ivoC_ |
|
not modelled |
63.6 |
21 |
PDB header:hydrolase
Chain: C: PDB Molecule:up1;
PDBTitle: structure of up1 protein
|
| 79 | c3cwcB_ |
|
not modelled |
63.6 |
23 |
PDB header:transferase
Chain: B: PDB Molecule:putative glycerate kinase 2;
PDBTitle: crystal structure of putative glycerate kinase 2 from salmonella2 typhimurium lt2
|
| 80 | c1xdiA_ |
|
not modelled |
63.4 |
27 |
PDB header:unknown function
Chain: A: PDB Molecule:rv3303c-lpda;
PDBTitle: crystal structure of lpda (rv3303c) from mycobacterium tuberculosis
|
| 81 | c3lwzC_ |
|
not modelled |
63.3 |
33 |
PDB header:lyase
Chain: C: PDB Molecule:3-dehydroquinate dehydratase;
PDBTitle: 1.65 angstrom resolution crystal structure of type ii 3-2 dehydroquinate dehydratase (aroq) from yersinia pestis
|
| 82 | c1fmtA_ |
|
not modelled |
63.2 |
21 |
PDB header:formyltransferase
Chain: A: PDB Molecule:methionyl-trna fmet formyltransferase;
PDBTitle: methionyl-trnafmet formyltransferase from escherichia coli
|
| 83 | c2a87A_ |
|
not modelled |
62.8 |
10 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:thioredoxin reductase;
PDBTitle: crystal structure of m. tuberculosis thioredoxin reductase
|
| 84 | c2ywjA_ |
|
not modelled |
62.5 |
16 |
PDB header:transferase
Chain: A: PDB Molecule:glutamine amidotransferase subunit pdxt;
PDBTitle: crystal structure of uncharacterized conserved protein from2 methanocaldococcus jannaschii
|
| 85 | c3q0iA_ |
|
not modelled |
62.4 |
17 |
PDB header:transferase
Chain: A: PDB Molecule:methionyl-trna formyltransferase;
PDBTitle: methionyl-trna formyltransferase from vibrio cholerae
|
| 86 | c2bcpA_ |
|
not modelled |
62.1 |
10 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:nadh oxidase;
PDBTitle: structural analysis of streptococcus pyogenes nadh oxidase:2 c44s nox with azide
|
| 87 | d1ihua1 |
|
not modelled |
61.8 |
20 |
Fold:P-loop containing nucleoside triphosphate hydrolases
Superfamily:P-loop containing nucleoside triphosphate hydrolases
Family:Nitrogenase iron protein-like |
| 88 | d1pjqa1 |
|
not modelled |
61.7 |
27 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Siroheme synthase N-terminal domain-like |
| 89 | c2eq8E_ |
|
not modelled |
61.5 |
17 |
PDB header:oxidoreductase
Chain: E: PDB Molecule:pyruvate dehydrogenase complex, dihydrolipoamide
PDBTitle: crystal structure of lipoamide dehydrogenase from thermus thermophilus2 hb8 with psbdp
|
| 90 | c3eagA_ |
|
not modelled |
61.0 |
21 |
PDB header:ligase
Chain: A: PDB Molecule:udp-n-acetylmuramate:l-alanyl-gamma-d-glutamyl-meso-
PDBTitle: the crystal structure of udp-n-acetylmuramate:l-alanyl-gamma-d-2 glutamyl-meso-diaminopimelate ligase (mpl) from neisseria3 meningitides
|
| 91 | c3c6kC_ |
|
not modelled |
61.0 |
19 |
PDB header:transferase
Chain: C: PDB Molecule:spermine synthase;
PDBTitle: crystal structure of human spermine synthase in complex2 with spermidine and 5-methylthioadenosine
|
| 92 | d2piaa2 |
|
not modelled |
61.0 |
21 |
Fold:Ferredoxin reductase-like, C-terminal NADP-linked domain
Superfamily:Ferredoxin reductase-like, C-terminal NADP-linked domain
Family:Aromatic dioxygenase reductase-like |
| 93 | c2xdoC_ |
|
not modelled |
60.9 |
19 |
PDB header:oxidoreductase
Chain: C: PDB Molecule:tetx2 protein;
PDBTitle: structure of the tetracycline degrading monooxygenase tetx2 from2 bacteroides thetaiotaomicron
|
| 94 | d1qfja2 |
|
not modelled |
60.8 |
21 |
Fold:Ferredoxin reductase-like, C-terminal NADP-linked domain
Superfamily:Ferredoxin reductase-like, C-terminal NADP-linked domain
Family:Reductases |
| 95 | d1cqxa3 |
|
not modelled |
60.0 |
24 |
Fold:Ferredoxin reductase-like, C-terminal NADP-linked domain
Superfamily:Ferredoxin reductase-like, C-terminal NADP-linked domain
Family:Flavohemoglobin, C-terminal domain |
| 96 | d1mo9a2 |
|
not modelled |
59.7 |
23 |
Fold:FAD/NAD(P)-binding domain
Superfamily:FAD/NAD(P)-binding domain
Family:FAD/NAD-linked reductases, N-terminal and central domains |
| 97 | d2gv8a1 |
|
not modelled |
59.7 |
12 |
Fold:FAD/NAD(P)-binding domain
Superfamily:FAD/NAD(P)-binding domain
Family:FAD/NAD-linked reductases, N-terminal and central domains |
| 98 | d1djqa3 |
|
not modelled |
59.5 |
19 |
Fold:Nucleotide-binding domain
Superfamily:Nucleotide-binding domain
Family:N-terminal domain of adrenodoxin reductase-like |
| 99 | d1umka2 |
|
not modelled |
59.3 |
17 |
Fold:Ferredoxin reductase-like, C-terminal NADP-linked domain
Superfamily:Ferredoxin reductase-like, C-terminal NADP-linked domain
Family:Reductases |
| 100 | d1hyha1 |
|
not modelled |
59.1 |
22 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:LDH N-terminal domain-like |
| 101 | c3ouzA_ |
|
not modelled |
59.1 |
11 |
PDB header:ligase
Chain: A: PDB Molecule:biotin carboxylase;
PDBTitle: crystal structure of biotin carboxylase-adp complex from campylobacter2 jejuni
|
| 102 | d2voua1 |
|
not modelled |
59.0 |
21 |
Fold:FAD/NAD(P)-binding domain
Superfamily:FAD/NAD(P)-binding domain
Family:FAD-linked reductases, N-terminal domain |
| 103 | c3dhnA_ |
|
not modelled |
58.9 |
22 |
PDB header:isomerase, lyase
Chain: A: PDB Molecule:nad-dependent epimerase/dehydratase;
PDBTitle: crystal structure of the putative epimerase q89z24_bactn2 from bacteroides thetaiotaomicron. northeast structural3 genomics consortium target btr310.
|
| 104 | c1f8sA_ |
|
not modelled |
58.8 |
20 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:l-amino acid oxidase;
PDBTitle: crystal structure of l-amino acid oxidase from calloselasma2 rhodostoma, complexed with three molecules of o-aminobenzoate.
|
| 105 | c3d8xB_ |
|
not modelled |
58.7 |
14 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:thioredoxin reductase 1;
PDBTitle: crystal structure of saccharomyces cerevisiae nadph dependent2 thioredoxin reductase 1
|
| 106 | c3bzbA_ |
|
not modelled |
58.7 |
23 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:uncharacterized protein;
PDBTitle: crystal structure of uncharacterized protein cmq451c from the2 primitive red alga cyanidioschyzon merolae
|
| 107 | d2iida1 |
|
not modelled |
58.6 |
20 |
Fold:FAD/NAD(P)-binding domain
Superfamily:FAD/NAD(P)-binding domain
Family:FAD-linked reductases, N-terminal domain |
| 108 | d1h6va2 |
|
not modelled |
58.5 |
14 |
Fold:FAD/NAD(P)-binding domain
Superfamily:FAD/NAD(P)-binding domain
Family:FAD/NAD-linked reductases, N-terminal and central domains |
| 109 | c3pvcA_ |
|
not modelled |
58.5 |
13 |
PDB header:oxidoreductase, transferase
Chain: A: PDB Molecule:trna 5-methylaminomethyl-2-thiouridine biosynthesis
PDBTitle: crystal structure of apo mnmc from yersinia pestis
|
| 110 | c3kipU_ |
|
not modelled |
58.4 |
28 |
PDB header:lyase
Chain: U: PDB Molecule:3-dehydroquinase, type ii;
PDBTitle: crystal structure of type-ii 3-dehydroquinase from c. albicans
|
| 111 | d1w4xa2 |
|
not modelled |
58.0 |
19 |
Fold:FAD/NAD(P)-binding domain
Superfamily:FAD/NAD(P)-binding domain
Family:FAD/NAD-linked reductases, N-terminal and central domains |
| 112 | c3lzxB_ |
|
not modelled |
58.0 |
12 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:ferredoxin--nadp reductase 2;
PDBTitle: crystal structure of ferredoxin-nadp+ oxidoreductase from bacillus2 subtilis (form ii)
|
| 113 | c3hn7A_ |
|
not modelled |
57.8 |
27 |
PDB header:ligase
Chain: A: PDB Molecule:udp-n-acetylmuramate-l-alanine ligase;
PDBTitle: crystal structure of a murein peptide ligase mpl (psyc_0032) from2 psychrobacter arcticus 273-4 at 1.65 a resolution
|
| 114 | c3nrnA_ |
|
not modelled |
57.7 |
24 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:uncharacterized protein pf1083;
PDBTitle: crystal structure of pf1083 protein from pyrococcus furiosus,2 northeast structural genomics consortium target pfr223
|
| 115 | c1vjtA_ |
|
not modelled |
57.7 |
19 |
PDB header:hydrolase
Chain: A: PDB Molecule:alpha-glucosidase;
PDBTitle: crystal structure of alpha-glucosidase (tm0752) from thermotoga2 maritima at 2.50 a resolution
|
| 116 | d1d7ya1 |
|
not modelled |
57.5 |
27 |
Fold:FAD/NAD(P)-binding domain
Superfamily:FAD/NAD(P)-binding domain
Family:FAD/NAD-linked reductases, N-terminal and central domains |
| 117 | d1s3ia2 |
|
not modelled |
57.3 |
21 |
Fold:Formyltransferase
Superfamily:Formyltransferase
Family:Formyltransferase |
| 118 | c1v59B_ |
|
not modelled |
57.0 |
24 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:dihydrolipoamide dehydrogenase;
PDBTitle: crystal structure of yeast lipoamide dehydrogenase2 complexed with nad+
|
| 119 | c2xagA_ |
|
not modelled |
56.9 |
23 |
PDB header:transcription
Chain: A: PDB Molecule:lysine-specific histone demethylase 1;
PDBTitle: crystal structure of lsd1-corest in complex with para-bromo-2 (-)-trans-2-phenylcyclopropyl-1-amine
|
| 120 | c2v1dA_ |
|
not modelled |
56.9 |
23 |
PDB header:oxidoreductase/repressor
Chain: A: PDB Molecule:lysine-specific histone demethylase 1;
PDBTitle: structural basis of lsd1-corest selectivity in histone h32 recognition
|